

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149207.4 - phase: 0
(168 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
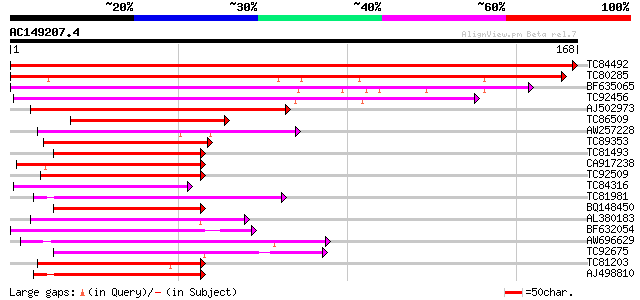
Score E
Sequences producing significant alignments: (bits) Value
TC84492 similar to GP|23498276|emb|CAD49247. erythrocyte membran... 327 2e-90
TC80285 similar to GP|7300092|gb|AAF55261.1| srp gene product {D... 145 1e-35
BF635065 weakly similar to GP|11037008|gb latent nuclear antigen... 128 9e-31
TC92456 weakly similar to PIR|T46707|T46707 proteophosphoglycan ... 120 2e-28
AJ502973 70 3e-13
TC86509 similar to GP|6361629|dbj|BAA86656.1 vitellogenin {Perip... 60 5e-10
AW257228 GP|17342711|gb 1-aminocyclopropanecarboxylic acid oxida... 50 3e-07
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 50 4e-07
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 50 6e-07
CA917238 weakly similar to PIR|A84538|A845 hypothetical protein ... 49 1e-06
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 47 5e-06
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 45 1e-05
TC81981 similar to GP|11994545|dbj|BAB02732. emb|CAB62106.1~gene... 45 1e-05
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 44 3e-05
AL380183 44 3e-05
BF632054 similar to GP|10177628|dbj gb|AAF30317.1~gene_id:K6M13.... 44 3e-05
AW696629 44 4e-05
TC92675 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 43 5e-05
TC81203 weakly similar to GP|10177464|dbj|BAB10855. gb|AAD21700.... 43 7e-05
AJ498810 weakly similar to PIR|D86339|D863 protein F2D10.14 [imp... 41 2e-04
>TC84492 similar to GP|23498276|emb|CAD49247. erythrocyte membrane protein 1
(PfEMP1) {Plasmodium falciparum 3D7}, partial (1%)
Length = 751
Score = 327 bits (837), Expect = 2e-90
Identities = 167/168 (99%), Positives = 167/168 (99%)
Frame = +3
Query: 1 MMADATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLA 60
MMADATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLA
Sbjct: 183 MMADATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLA 362
Query: 61 DITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELD 120
DITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELD
Sbjct: 363 DITDLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELD 542
Query: 121 NLLVGVRSIKRELEIIKVDTLAMKDRMKCLESFLKIYLKSATSSSSQL 168
NLLVGVRSIKRELEIIKVDTLAMKDRMKCLESFLKIYLKS TSSSSQL
Sbjct: 543 NLLVGVRSIKRELEIIKVDTLAMKDRMKCLESFLKIYLKSTTSSSSQL 686
>TC80285 similar to GP|7300092|gb|AAF55261.1| srp gene product {Drosophila
melanogaster}, partial (2%)
Length = 1055
Score = 145 bits (365), Expect = 1e-35
Identities = 86/185 (46%), Positives = 118/185 (63%), Gaps = 20/185 (10%)
Frame = -2
Query: 1 MMADATAESP-TTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSL 59
M A+ATAE P T +LP EL+IEI++RVDS N LQLR VCK W SL+ D +FV+KH
Sbjct: 1039 MTAEATAELPKTVVLPEELIIEIISRVDSRNTLQLRPVCKRWNSLITDPEFVKKHFQSLF 860
Query: 60 ADITDLLSTATEHRKSFVS-HQIPNQL--QEEEEEDDDDDDDDEEV------------DE 104
DI DL S A E +F+S H I N + QEE EE+ +++++DE+V +E
Sbjct: 859 TDIRDLTSKALEQSNAFISQHNINNPVVPQEEGEENAEEEEEDEDVAFVDEEVVAAVDEE 680
Query: 105 EEAAKRARMNRLAELDNLLVGVRSIKRELEIIKVD----TLAMKDRMKCLESFLKIYLKS 160
++ KR M+ ++++DNLLV +R IK E I VD + A++D MKC SF++IYLK
Sbjct: 679 KDELKRVLMDLVSDMDNLLVNLRFIKDNTETIHVDMQTQSQALEDIMKCFRSFVRIYLK* 500
Query: 161 ATSSS 165
TSSS
Sbjct: 499 ETSSS 485
>BF635065 weakly similar to GP|11037008|gb latent nuclear antigen {Human
herpesvirus 8}, partial (3%)
Length = 681
Score = 128 bits (322), Expect = 9e-31
Identities = 79/197 (40%), Positives = 114/197 (57%), Gaps = 42/197 (21%)
Frame = +2
Query: 1 MMADATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLA 60
MM +ATAESP+++LP +LMIEIL+RV+SNNPL+LR VCK W SL++D QF+ HL +
Sbjct: 89 MMEEATAESPSSVLPEDLMIEILSRVESNNPLELRCVCKLWNSLILDSQFMMNHLDRLYT 268
Query: 61 DITDLLSTATEHRKSFVSHQIPNQ--LQEEEEEDDDDDD--DDEEVDE----EEAA---- 108
++T L + A EH +F S I N + +E+E DDDDDD DDEE DE EEAA
Sbjct: 269 EMTVLYAKAMEHLMAFKSQHIVNNPVIPQEQEHDDDDDDVEDDEETDEEDDNEEAAVEQE 448
Query: 109 -----KRARMNRLAELDNL--------------------LVGVRSIKRELEIIKVD---- 139
K+ MN L ++DN+ L +R ++ ++E K++
Sbjct: 449 EQKEEKQVTMNELVQMDNVDEVDKEKQLAKDVVAQLDYQLEMLRFVREDVETKKINIDMQ 628
Query: 140 -TLAMKDRMKCLESFLK 155
+ ++DRM L SF++
Sbjct: 629 PNIDLEDRMTYLRSFIR 679
>TC92456 weakly similar to PIR|T46707|T46707 proteophosphoglycan
membrane-associated [imported] - Leishmania major
(fragment), partial (10%)
Length = 538
Score = 120 bits (302), Expect = 2e-28
Identities = 74/157 (47%), Positives = 93/157 (59%), Gaps = 19/157 (12%)
Frame = -1
Query: 2 MADATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLAD 61
MA+AT ESPTTILP E+MIEIL RV+ N+ LQLR VCK WKSLV D QFV+ HL K L D
Sbjct: 472 MAEATVESPTTILPEEVMIEILYRVELNSTLQLRCVCKLWKSLVHDSQFVKNHLFKLLND 293
Query: 62 ITDLLSTATEHRKSFVSHQIPN-----QLQEEEEEDDDDDDDDEEVD------------- 103
IT L S E +F S + N Q QE+E+ D+D +D + D
Sbjct: 292 ITVLFSKTAEQFNAFKSQHLINNPVVPQEQEQEQVVDEDGEDAAQEDDNAAAEEGDAADE 113
Query: 104 -EEEAAKRARMNRLAELDNLLVGVRSIKRELEIIKVD 139
EEE K M LA+LD +LV V+S+K + + I +D
Sbjct: 112 EEEEDEKHWLMTALAKLDGVLVNVQSLKGDFKFINLD 2
>AJ502973
Length = 338
Score = 70.5 bits (171), Expect = 3e-13
Identities = 35/77 (45%), Positives = 50/77 (64%)
Frame = +3
Query: 7 AESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDLL 66
A SP ++LP EL++EIL+RVDS+N L LR VC WKSL+ D QF++ H+ +S+ + L
Sbjct: 87 AVSPISVLPDELLMEILSRVDSSNHLDLRCVCNLWKSLIRDPQFMKNHILRSITGFSSLS 266
Query: 67 STATEHRKSFVSHQIPN 83
H +F SH + N
Sbjct: 267 YKIEGHFIAFKSHIVYN 317
>TC86509 similar to GP|6361629|dbj|BAA86656.1 vitellogenin {Periplaneta
americana}, partial (1%)
Length = 905
Score = 59.7 bits (143), Expect = 5e-10
Identities = 25/47 (53%), Positives = 36/47 (76%)
Frame = -3
Query: 19 MIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDL 65
++ I++R++SNNPL LR +CK WKSLVVD FV+ HL + L D +D+
Sbjct: 717 LLLIMSRINSNNPLDLRCICKLWKSLVVDHVFVRNHLSRMLTDFSDV 577
>AW257228 GP|17342711|gb 1-aminocyclopropanecarboxylic acid oxidase {Medicago
truncatula}, partial (11%)
Length = 702
Score = 50.4 bits (119), Expect = 3e-07
Identities = 31/84 (36%), Positives = 49/84 (57%), Gaps = 6/84 (7%)
Frame = +1
Query: 9 SPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQ-FVQKHLHKS-----LADI 62
+PT++L EL++EIL+R+ +Q + VCK WK+L+ DD F + HLH+S LA +
Sbjct: 331 APTSLLLDELIVEILSRLPVKTLMQFKCVCKSWKTLISDDPVFAKFHLHRSPRNTHLAIL 510
Query: 63 TDLLSTATEHRKSFVSHQIPNQLQ 86
+D T E S V + + L+
Sbjct: 511 SDRSITEDETDCSVVPFPVTHLLE 582
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 50.1 bits (118), Expect = 4e-07
Identities = 23/50 (46%), Positives = 33/50 (66%)
Frame = +3
Query: 11 TTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLA 60
T +P E+++EIL R+ + LQ R VCK WK+L+ D QF +KH+ S A
Sbjct: 96 TADMPEEIIVEILLRLPVRSLLQFRCVCKLWKTLISDPQFAKKHVSISTA 245
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported]
- Arabidopsis thaliana, partial (9%)
Length = 728
Score = 49.7 bits (117), Expect = 6e-07
Identities = 22/45 (48%), Positives = 30/45 (65%)
Frame = +1
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
LP E+ EIL+RV + L+LRS CKWW++L+ F+ HL KS
Sbjct: 16 LPTEVTTEILSRVPAKPLLRLRSTCKWWRNLIDSTDFIFLHLSKS 150
>CA917238 weakly similar to PIR|A84538|A845 hypothetical protein At2g16220
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 48.5 bits (114), Expect = 1e-06
Identities = 22/57 (38%), Positives = 41/57 (71%), Gaps = 1/57 (1%)
Frame = +2
Query: 3 ADATAES-PTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
A+A AE+ P ++LP EL+ E+L+R D + +++R V ++W S++ D +FV+ H+ +S
Sbjct: 110 AEAQAEAEPPSVLPDELITEVLSRGDVKSLMRMRCVSEYWNSMISDPRFVKLHMKRS 280
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 46.6 bits (109), Expect = 5e-06
Identities = 23/49 (46%), Positives = 30/49 (60%)
Frame = +3
Query: 10 PTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
P LP EL+ EIL R+ QLR +CK + SL+ D +F +KHLH S
Sbjct: 162 PLPTLPFELVAEILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHLHSS 308
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 45.4 bits (106), Expect = 1e-05
Identities = 21/53 (39%), Positives = 30/53 (55%)
Frame = +3
Query: 2 MADATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKH 54
+A+ TA P LP EL++ IL R+ + L+ + VCK WK+L D F H
Sbjct: 6 VAETTANKPLPFLPEELIVIILLRLPVRSLLRFKCVCKSWKTLFSDTHFANNH 164
>TC81981 similar to GP|11994545|dbj|BAB02732.
emb|CAB62106.1~gene_id:MGD8.12~similar to unknown
protein {Arabidopsis thaliana}, partial (4%)
Length = 635
Score = 45.1 bits (105), Expect = 1e-05
Identities = 27/75 (36%), Positives = 42/75 (56%)
Frame = +2
Query: 8 ESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDLLS 67
E PT LP +L+ EIL R+ +QLR +C+++ SL+ + +F +KHL S L++
Sbjct: 56 ELPT--LPFDLLPEILCRLPVKLLVQLRCLCEFFNSLISNPKFAKKHLQLSTKRHHLLVT 229
Query: 68 TATEHRKSFVSHQIP 82
+ R FV H P
Sbjct: 230 SWNISRGEFVQHDFP 274
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar
to unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 43.9 bits (102), Expect = 3e-05
Identities = 20/45 (44%), Positives = 28/45 (61%)
Frame = +3
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
LP E+ +EIL+R+ +Q + VCK WKS + FV+KHL S
Sbjct: 3 LPFEIQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKHLRVS 137
>AL380183
Length = 467
Score = 43.9 bits (102), Expect = 3e-05
Identities = 26/72 (36%), Positives = 41/72 (56%), Gaps = 7/72 (9%)
Frame = +1
Query: 7 AESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHL-------HKSL 59
AE+ LP++++ EIL R+ QLR +CK + SL+ D +F +KHL H L
Sbjct: 61 AETQLPTLPIDVLPEILCRLPIKLLGQLRCLCKSFNSLISDPKFAKKHLQLSTKRHHLML 240
Query: 60 ADITDLLSTATE 71
I D +S+++E
Sbjct: 241 TCIRDQISSSSE 276
>BF632054 similar to GP|10177628|dbj gb|AAF30317.1~gene_id:K6M13.17~similar
to unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 462
Score = 43.9 bits (102), Expect = 3e-05
Identities = 25/73 (34%), Positives = 38/73 (51%)
Frame = +3
Query: 1 MMADATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLA 60
M ++T P LP EL++EIL ++ + L+ R VCK W ++ + F++K LH S
Sbjct: 105 MNQNSTMILPLPFLPEELILEILIKLPIKSLLRFRCVCKSWLHIISNPYFIKKQLHFS-- 278
Query: 61 DITDLLSTATEHR 73
T T HR
Sbjct: 279 --TQNTHFTTNHR 311
>AW696629
Length = 663
Score = 43.5 bits (101), Expect = 4e-05
Identities = 27/94 (28%), Positives = 52/94 (54%), Gaps = 2/94 (2%)
Frame = +2
Query: 4 DATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADIT 63
+AT+ P ILP +L+++IL+ + ++ SV K WKSL++D F + HL KS +
Sbjct: 41 EATSSPP--ILPSDLIMQILSWLPVKLLIRFTSVSKHWKSLILDPNFAKLHLQKSPKNTH 214
Query: 64 DLLSTATEHRKSFV--SHQIPNQLQEEEEEDDDD 95
+L+ + ++V + + + L E+ D++
Sbjct: 215 MILTALDDEDDTWVVTPYPVRSLLLEQSSFSDEE 316
>TC92675 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (17%)
Length = 630
Score = 43.1 bits (100), Expect = 5e-05
Identities = 29/83 (34%), Positives = 42/83 (49%), Gaps = 2/83 (2%)
Frame = +3
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLH--KSLADITDLLSTATE 71
+P ELMI+IL R+ + ++ +SVCK W SLV D F H + + +L +T
Sbjct: 72 MPNELMIQILLRLPVKSLIRFKSVCKSWFSLVSDTHFAYSHFNFPPQHTLVGRVLFISTS 251
Query: 72 HRKSFVSHQIPNQLQEEEEEDDD 94
RK S I L+ +DDD
Sbjct: 252 ARK---SRSIDLDLEASPLDDDD 311
>TC81203 weakly similar to GP|10177464|dbj|BAB10855.
gb|AAD21700.1~gene_id:MQB2.18~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 715
Score = 42.7 bits (99), Expect = 7e-05
Identities = 20/51 (39%), Positives = 34/51 (66%), Gaps = 1/51 (1%)
Frame = +1
Query: 9 SPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVV-DDQFVQKHLHKS 58
+PT++L EL+++IL+R+ +Q + VCK WK+L+ D F + HL +S
Sbjct: 184 APTSLLLDELIVDILSRLPVKTLMQFKCVCKSWKTLISHDPSFAKLHLQRS 336
>AJ498810 weakly similar to PIR|D86339|D863 protein F2D10.14 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 715
Score = 41.2 bits (95), Expect = 2e-04
Identities = 22/51 (43%), Positives = 32/51 (62%)
Frame = +3
Query: 8 ESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
E PT LP +++ EIL R+ +QLR +CK++ SL+ D F +KHL S
Sbjct: 258 ELPT--LPFDVLPEILCRLPVKLLIQLRCLCKFFNSLISDPTFAKKHLQLS 404
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.129 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,448,125
Number of Sequences: 36976
Number of extensions: 59180
Number of successful extensions: 3170
Number of sequences better than 10.0: 327
Number of HSP's better than 10.0 without gapping: 1038
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2009
length of query: 168
length of database: 9,014,727
effective HSP length: 89
effective length of query: 79
effective length of database: 5,723,863
effective search space: 452185177
effective search space used: 452185177
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC149207.4


