

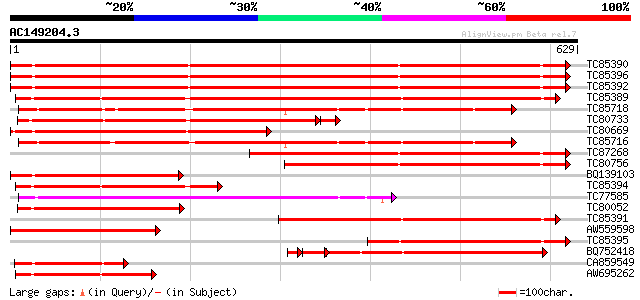
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149204.3 - phase: 0
(629 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 763 0.0
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 761 0.0
TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chape... 757 0.0
TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protei... 578 e-165
TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa prot... 435 e-122
TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa prot... 421 e-122
TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 411 e-115
TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chape... 410 e-114
TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chape... 369 e-102
TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chape... 320 1e-87
BQ139103 homologue to GP|2655420|gb| heat shock cognate protein ... 283 1e-76
TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic retic... 277 8e-75
TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock... 252 3e-67
TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chapero... 250 1e-66
TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chape... 249 3e-66
AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 ... 234 7e-62
TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protei... 205 4e-53
BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated pr... 187 2e-51
CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus sto... 169 4e-42
AW695262 homologue to GP|170386|gb|A glucose-regulated protein 7... 167 1e-41
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 763 bits (1970), Expect = 0.0
Identities = 400/625 (64%), Positives = 490/625 (78%), Gaps = 3/625 (0%)
Frame = +1
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS VAFTD+ RLIG+AA
Sbjct: 136 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 309
Query: 61 KNQAASNPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCF 120
KNQ A NPTNT+FDAKRLIGR+F D ++ D+KLWPFKVIPG D P I+V YK +EK F
Sbjct: 310 KNQVAMNPTNTVFDAKRLIGRRFGDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 489
Query: 121 VAEEISSMILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIIN 180
AEEISSM+L KM+EIAE +L +KNAV+TVPAYFNDSQR+ATKDAG+I+GLNV+RIIN
Sbjct: 490 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVLRIIN 669
Query: 181 EPTAAALAYGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGE 240
EPTAAA+AYG+ K+ G++NV IFDLGGGTFDVSLLT+++ FEVKATAGDTHLGGE
Sbjct: 670 EPTAAAIAYGLDKKATST-GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGE 846
Query: 241 DFDNKMVSHFVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEATIDIDAICQGI 300
DFDN+MV+HFV E KRKN DIS NP+ALR+LRTACERAKR LS + TI+ID++ +GI
Sbjct: 847 DFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGI 1026
Query: 301 DFCSSITRAKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQ 360
DF ++ITRA+FE LNMDLF KCME V CLRDA MD++ VH++VLVGGS+RIPKV++LLQ
Sbjct: 1027DFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGSTRIPKVQQLLQ 1206
Query: 361 DFFKGRDLCMSINPDEAVAYGAAVQAALLI-DGIKNVPNLVLRDITPLSLGIWTKGDVMS 419
DFF G++LC SINPDEAVAYGAAVQAA+L +G + V +L+L D+TPLSLG+ T G VM+
Sbjct: 1207DFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMT 1386
Query: 420 VVIPKNTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERIKASENNLLGLFRLF-VPPAPRG 478
V+IP+NT IP K E T DNQ GV I VYEGER + +NNLLG F L +PPAPRG
Sbjct: 1387VLIPRNTTIPTK-KEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRG 1563
Query: 479 L-CIKVCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEAEKFKVEDM 537
+ I VCF IDA+GILNV+AE++TTG K++ITITND RLS +EIE+M+QEAEK+K ED
Sbjct: 1564VPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDE 1743
Query: 538 KFQEKARAVNALEDYLYKTSKVMEDNSVSSMLAPADKMKIDSAIMNGKSLIDNHGNQHNE 597
+ ++K A NALE+Y Y ++D+ ++S L+ DK KI+ AI +D GNQ E
Sbjct: 1744EHKKKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLD--GNQLGE 1917
Query: 598 TYVFVDFLKELEIIFESSLNKINKG 622
F D +KELE I + ++ +G
Sbjct: 1918ADEFEDKMKELEGICNPIIARMYQG 1992
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 761 bits (1964), Expect = 0.0
Identities = 399/625 (63%), Positives = 489/625 (77%), Gaps = 3/625 (0%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS VAFTDS RLIG+AA
Sbjct: 107 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 280
Query: 61 KNQAASNPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCF 120
KNQ A NPTNT+FDAKRLIGR+ SD+ ++ D+KLWPFKVI G + P I V YKG+EK F
Sbjct: 281 KNQVAMNPTNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEEKLF 460
Query: 121 VAEEISSMILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIIN 180
+EEISSM+L KM+EIAE +L + +KNAV+TVPAYFNDSQR+ATKDAG+IAGLNV+RIIN
Sbjct: 461 ASEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 640
Query: 181 EPTAAALAYGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGE 240
EPTAAA+AYG+ K+ V G++NV IFDLGGGTFDVSLLT+++ FEVKATAGDTHLGGE
Sbjct: 641 EPTAAAIAYGLDKKATSV-GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGE 817
Query: 241 DFDNKMVSHFVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEATIDIDAICQGI 300
DFDN+MV+HFV E KRKN DIS NP+ALR+LRTACERAKR LS + TI+ID++ +G+
Sbjct: 818 DFDNRMVTHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGV 997
Query: 301 DFCSSITRAKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQ 360
DF ++ITRA+FE LNMDLF KCME V CLRDA MD+S VH++VLVGGS+RIPKV++LLQ
Sbjct: 998 DFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQ 1177
Query: 361 DFFKGRDLCMSINPDEAVAYGAAVQAALLI-DGIKNVPNLVLRDITPLSLGIWTKGDVMS 419
DFF G++LC SINPDEAVAYGAAVQAA+L +G + V +L+L D+TPLS G+ T G VM+
Sbjct: 1178DFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSQGLETAGGVMT 1357
Query: 420 VVIPKNTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERIKASENNLLGLFRLF-VPPAPRG 478
V+IP+NT IP K E T DNQ GV I VYEGER + +NNLLG F L +PPAPRG
Sbjct: 1358VLIPRNTTIPTK-KEQVFSTYSDNQPGVLIQVYEGERTRTKDNNLLGKFELSGIPPAPRG 1534
Query: 479 L-CIKVCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEAEKFKVEDM 537
+ I VCF IDA+GILNV+AE++TTG K++ITITND RLS +EIE+M+QEAEK+K ED
Sbjct: 1535VPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDE 1714
Query: 538 KFQEKARAVNALEDYLYKTSKVMEDNSVSSMLAPADKMKIDSAIMNGKSLIDNHGNQHNE 597
+ ++K A N+LE+Y Y ++D +SS L+ DK +I+ AI +D NQ E
Sbjct: 1715EHKKKVEAKNSLENYAYNMRNTIKDEKISSKLSGGDKKQIEDAIEGAIQWLD--ANQLAE 1888
Query: 598 TYVFVDFLKELEIIFESSLNKINKG 622
F D +KELE I + K+ +G
Sbjct: 1889ADEFEDKMKELETICNPIIAKMYQG 1963
>TC85392 homologue to PIR|JC4786|JC4786 dnaK-type molecular chaperone
hsc70-3 - tomato, complete
Length = 2295
Score = 757 bits (1955), Expect = 0.0
Identities = 398/625 (63%), Positives = 488/625 (77%), Gaps = 3/625 (0%)
Frame = +1
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS VAFTD+ RLIG+AA
Sbjct: 130 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 303
Query: 61 KNQAASNPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCF 120
KNQ A NPTNT+FDAKRLIGR+ SD ++ D+KLWPFKVIPG D P I+V YK +EK F
Sbjct: 304 KNQVAMNPTNTVFDAKRLIGRRISDVSVQSDMKLWPFKVIPGPADKPMIVVNYKAEEKQF 483
Query: 121 VAEEISSMILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIIN 180
AEEISSM+L KM+EIAE +L +KNAV+TVPAYFNDSQR+ATKDAG+I+GLNVMRIIN
Sbjct: 484 SAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 663
Query: 181 EPTAAALAYGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGE 240
EPTAAA+AYG+ K+ G++NV IFDLGGGTFDVSLLT+++ FEVKATAGDTHLGGE
Sbjct: 664 EPTAAAIAYGLDKKATST-GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGE 840
Query: 241 DFDNKMVSHFVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEATIDIDAICQGI 300
DFDN+MV+HFV E KRKN DIS NP+ALR++RTACERAKR LS + TI+ID++ +GI
Sbjct: 841 DFDNRMVNHFVQEFKRKNKKDISGNPRALRRVRTACERAKRTLSSTAQTTIEIDSLFEGI 1020
Query: 301 DFCSSITRAKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQ 360
DF ++ITRA+FE LNMDLF KCME V CLRDA MD++ V ++VLVGGS+RIPKV++LLQ
Sbjct: 1021DFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVDDVVLVGGSTRIPKVQQLLQ 1200
Query: 361 DFFKGRDLCMSINPDEAVAYGAAVQAALLI-DGIKNVPNLVLRDITPLSLGIWTKGDVMS 419
DFF G++LC SINPDEAVAYGAAVQAA+L +G + V +L+L D+TPLSLG+ T G VM+
Sbjct: 1201DFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMT 1380
Query: 420 VVIPKNTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERIKASENNLLGLFRLF-VPPAPRG 478
V+IP+NT IP K E T DNQ GV I VYEGER + +NNLLG F L +PPAPRG
Sbjct: 1381VLIPRNTTIPTK-KEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRG 1557
Query: 479 L-CIKVCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEAEKFKVEDM 537
+ I VCF IDA+GILNV+AE++TTG K++ITITND RLS ++IE+M+QEAEK+K ED
Sbjct: 1558VPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDE 1737
Query: 538 KFQEKARAVNALEDYLYKTSKVMEDNSVSSMLAPADKMKIDSAIMNGKSLIDNHGNQHNE 597
+ + K A NALE+Y Y ++D+ ++S L+ DK KI+ AI +D GNQ E
Sbjct: 1738EHKRKVEAKNALENYAYNMRNTIKDDKIASKLSADDKKKIEDAIEGAIQWLD--GNQLGE 1911
Query: 598 TYVFVDFLKELEIIFESSLNKINKG 622
F D +KELE I + ++ +G
Sbjct: 1912ADEFEDKMKELEGICNPIIARMYQG 1986
>TC85389 homologue to SP|Q03684|BIP4_TOBAC Luminal binding protein 4
precursor (BiP 4) (78 kDa glucose-regulated protein
homolog 4) (GRP 78-4)., partial (94%)
Length = 2265
Score = 578 bits (1489), Expect = e-165
Identities = 316/610 (51%), Positives = 434/610 (70%), Gaps = 5/610 (0%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAAKNQAAS 66
G IGIDLGTTYSCV V+ +N EII NDQGNR TPS V+FTD RLIG AAKN AA
Sbjct: 155 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 328
Query: 67 NPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYK-GKEKCFVAEEI 125
NP TIFD KRLIGRKF+D +++D+KL P+K++ D P I V+ K G+ K F EE+
Sbjct: 329 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIV-NKDGKPYIQVRVKDGETKVFSPEEV 505
Query: 126 SSMILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIINEPTAA 185
S+MIL+KM+E AE FL +++AV+TVPAYFND+QR+ATKDAG+IAGLNV RIINEPTAA
Sbjct: 506 SAMILTKMKETAEAFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 685
Query: 186 ALAYGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGEDFDNK 245
A+AYG+ K+G G++N+ +FDLGGGTFDVS+LT+ + FEV +T GDTHLGGEDFD +
Sbjct: 686 AIAYGLDKKG----GEKNILVFDLGGGTFDVSILTIDNGVFEVLSTNGDTHLGGEDFDQR 853
Query: 246 MVSHFVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEATIDIDAICQGIDFCSS 305
++ +F+ +K+K++ DIS + +AL KLR ERAKRALS + ++I+++ G+DF
Sbjct: 854 IMEYFIKLIKKKHSKDISKDNRALGKLRRESERAKRALSSQHQVRVEIESLFDGVDFSEP 1033
Query: 306 ITRAKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQDFFKG 365
+TRA+FE LN DLF K M V+ + DA + ++ + EIVLVGGS+RIPKV++LL+D+F G
Sbjct: 1034LTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDG 1213
Query: 366 RDLCMSINPDEAVAYGAAVQAALLI-DGIKNVPNLVLRDITPLSLGIWTKGDVMSVVIPK 424
++ +NPDEAVA+GAAVQ ++L +G + +++L D+ PL+LGI T G VM+ +IP+
Sbjct: 1214KEPNKGVNPDEAVAFGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPR 1393
Query: 425 NTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERIKASENNLLGLFRLF-VPPAPRGL-CIK 482
NT IP K ++ T D Q VSI V+EGER + LLG F L +PPAPRG I+
Sbjct: 1394NTVIPTKKSQ-VFTTYQDQQTTVSIQVFEGERSLTKDCRLLGNFDLSGIPPAPRGTPQIE 1570
Query: 483 VCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEAEKFKVEDMKFQEK 542
V F +DA+GILNV AE++ TG +ITITN+ RLS +EI+RM++EAE+F ED K +E+
Sbjct: 1571VTFEVDANGILNVRAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKER 1750
Query: 543 ARAVNALEDYLYK-TSKVMEDNSVSSMLAPADKMKIDSAIMNGKSLIDNHGNQHNETYVF 601
A NALE Y+Y +++ + + ++ L +K KI++A+ +D+ NQ E F
Sbjct: 1751IDARNALETYVYNMKNQISDKDKLADKLESDEKEKIEAAVKEALEWLDD--NQTVEKEEF 1924
Query: 602 VDFLKELEII 611
+ LKE+E +
Sbjct: 1925EEKLKEVEAV 1954
>TC85718 homologue to SP|Q01899|HS7M_PHAVU Heat shock 70 kDa protein
mitochondrial precursor. [Kidney bean French bean]
{Phaseolus vulgaris}, complete
Length = 2371
Score = 435 bits (1118), Expect = e-122
Identities = 255/560 (45%), Positives = 362/560 (64%), Gaps = 7/560 (1%)
Frame = +2
Query: 10 IGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRL-IGNAAKNQAASNP 68
IGIDLGTT SCV+V + +N + ++ N +G RTTPSVVAFT L +G AK QA +NP
Sbjct: 263 IGIDLGTTNSCVSVMEGKNPK--VVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNP 436
Query: 69 TNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCFVAEEISSM 128
NTI AKRLIGR+F D +K++K+ P+K++ N V+ KG++ + +I +
Sbjct: 437 ENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAP--NGDAWVEAKGQQ--YSPSQIGAF 604
Query: 129 ILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIINEPTAAALA 188
+L+KM+E AE +L V AVITVPAYFND+QR+ATKDAG IAGL V+RIINEPTAAAL+
Sbjct: 605 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 784
Query: 189 YGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGEDFDNKMVS 248
YG+ K G + +FDLGGGTFDVS+L + + FEVKAT GDT LGGEDFDN ++
Sbjct: 785 YGMNKEG-------LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLD 943
Query: 249 HFVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEATIDIDAICQGIDFCS---- 304
VNE KR ++D+S + AL++LR A E+AK LS + I++ I
Sbjct: 944 FLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNI 1123
Query: 305 SITRAKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQDFFK 364
++TR+KFE L +L E+ +SCL+DA + + E++LVGG +R+PKV++++ F
Sbjct: 1124TLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF- 1300
Query: 365 GRDLCMSINPDEAVAYGAAVQAALLIDGIKNVPNLVLRDITPLSLGIWTKGDVMSVVIPK 424
G+ C +NPDEAVA GAA+Q +L +K L+L D+TPLSLGI T G + + +I +
Sbjct: 1301GKSPCKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLINR 1471
Query: 425 NTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERIKASENNLLGLFRLF-VPPAPRGL-CIK 482
NT IP K ++ T+ DNQ V I V +GER AS+N +LG F L +PPAPRG+ I+
Sbjct: 1472NTTIPTKKSQ-VFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIE 1648
Query: 483 VCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEAEKFKVEDMKFQEK 542
V F IDA+GI+ V+A++++TG + +ITI + LS EI+ M++EAE +D + +
Sbjct: 1649VTFDIDANGIVTVSAKDKSTGKEQQITIKSSG-GLSDDEIQNMVKEAELHAQKDQERKSL 1825
Query: 543 ARAVNALEDYLYKTSKVMED 562
N+ + +Y K + +
Sbjct: 1826IDIRNSADTSIYSIEKSLSE 1885
>TC80733 homologue to SP|Q01233|HS70_NEUCR Heat shock 70 kDa protein
(HSP70). {Neurospora crassa}, partial (54%)
Length = 1258
Score = 421 bits (1083), Expect(2) = e-122
Identities = 220/336 (65%), Positives = 267/336 (78%)
Frame = +2
Query: 9 AIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAAKNQAASNP 68
AIGIDLGTTYSCV + N++ EII NDQGNRTTPS VAF D+ RLIG+AAKNQ A NP
Sbjct: 188 AIGIDLGTTYSCVG--RYANDKIEIIANDQGNRTTPSYVAFNDTERLIGDAAKNQVAMNP 361
Query: 69 TNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCFVAEEISSM 128
NT+FDAKRLIGRKFSDS ++ D+K +PFKVI P I V++KG+ K F EEIS+M
Sbjct: 362 HNTVFDAKRLIGRKFSDSEVQADMKHFPFKVIDKGG-KPNIEVEFKGENKTFTPEEISAM 538
Query: 129 ILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIINEPTAAALA 188
+L KM+E AE +L V NAVITVPAYFNDSQR+ATKDAG+IAGLNV+RIINEPTAAA+A
Sbjct: 539 VLVKMRETAEAYLGGQVTNAVITVPAYFNDSQRQATKDAGLIAGLNVLRIINEPTAAAIA 718
Query: 189 YGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGEDFDNKMVS 248
YG+ K+ G+RNV IFDLGGGTFDVSLLT+++ FEVK+TAGDTHLGGEDFDN++V+
Sbjct: 719 YGLDKKAE---GERNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGEDFDNRLVN 889
Query: 249 HFVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEATIDIDAICQGIDFCSSITR 308
HFVNE KRK+ D+S N +ALR+LRTACERAKR LS + +I+ID++ +GIDF +SITR
Sbjct: 890 HFVNEFKRKHKKDLSSNARALRRLRTACERAKRTLSSSAQTSIEIDSLFEGIDFYTSITR 1069
Query: 309 AKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIV 344
A+FE L DLF ++ V L DA +D+S VHEIV
Sbjct: 1070ARFEELCQDLFRSTIQPVDRVLSDAKIDKSQVHEIV 1177
Score = 35.0 bits (79), Expect(2) = e-122
Identities = 12/22 (54%), Positives = 20/22 (90%)
Frame = +3
Query: 346 VGGSSRIPKVRKLLQDFFKGRD 367
VGGS+RIP+++KL+ D+F G++
Sbjct: 1182 VGGSTRIPRIQKLISDYFNGKE 1247
>TC80669 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (44%)
Length = 955
Score = 411 bits (1057), Expect = e-115
Identities = 209/290 (72%), Positives = 244/290 (84%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAA 60
MA K EG AIGIDLGTTYSCV VWQ N+R EII NDQGNRTTPS VAFTD+ RLIG+AA
Sbjct: 98 MATK-EGKAIGIDLGTTYSCVGVWQ--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 268
Query: 61 KNQAASNPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCF 120
KNQ A NP NT+FDAKRLIGR+FSD ++ D+KLWPFKV+PG + P I+V YKG+EK F
Sbjct: 269 KNQVAMNPQNTVFDAKRLIGRRFSDESVQNDMKLWPFKVVPGPAEKPMIVVNYKGEEKKF 448
Query: 121 VAEEISSMILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIIN 180
AEEISSM+L KM+E+AE FL PVKNAV+TVPAYFNDSQR+ATKDAG I+GLNV+RIIN
Sbjct: 449 AAEEISSMVLIKMREVAEAFLGHPVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 628
Query: 181 EPTAAALAYGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGE 240
EPTAAA+AYG+ K+ + G++NV IFDLGGGTFDVSLLT+++ FEVKATAGDTHLGGE
Sbjct: 629 EPTAAAIAYGLDKKAS-RKGEQNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGE 805
Query: 241 DFDNKMVSHFVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEAT 290
DFDN+MV+HFV+E +RKN DIS N +ALR+LRTACERAKR LS + T
Sbjct: 806 DFDNRMVNHFVSEFRRKNKKDISGNARALRRLRTACERAKRTLSSTAQTT 955
>TC85716 homologue to PIR|S19140|S19140 dnaK-type molecular chaperone PHSP1
precursor mitochondrial - garden pea, partial (98%)
Length = 2422
Score = 410 bits (1053), Expect = e-114
Identities = 242/562 (43%), Positives = 354/562 (62%), Gaps = 9/562 (1%)
Frame = +1
Query: 10 IGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRL-IGNAAKNQAASNP 68
IGIDLGTT SCV++ + +N + +I N +G RTTPSVVAF L +G AK QA +NP
Sbjct: 247 IGIDLGTTNSCVSLMEGKNPK--VIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNP 420
Query: 69 TNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCFVAEEISSM 128
TNT+F KRLIGR+F D +K++K+ P+K++ + + + + ++ + +I +
Sbjct: 421 TNTLFGTKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEIN----KQQYSPSQIGAF 588
Query: 129 ILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIINEPTAAALA 188
+L+KM+E AE +L + AV+TVPAYFND+QR+ATKDAG IAGL V RIINEPTAAAL+
Sbjct: 589 VLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQATKDAGRIAGLEVKRIINEPTAAALS 768
Query: 189 YGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGEDFDNKMVS 248
YG+ + + +FDLGGGTFDVS+L + + FEVKAT GDT LGGEDFDN ++
Sbjct: 769 YGMNNKEGLIA------VFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLD 930
Query: 249 HFVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEATIDIDAICQGIDFCS---- 304
V+E KR +++D++ + AL++LR A E+AK LS + I++ I
Sbjct: 931 FLVSEFKRTDSIDLAKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNI 1110
Query: 305 SITRAKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQDFFK 364
++TR+KFE L +L E+ +SCL+DA + V E++LVGG +R+PKV++++ + F
Sbjct: 1111TLTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQEVVSEIF- 1287
Query: 365 GRDLCMSINPDEAVAYGAAVQAALLIDGIKNVPNLVLRDITPLSLGIWTKGDVMSVVIPK 424
G+ +NPDEAVA GAA+Q +L +K L+L D+TPLSLGI T G + + +I +
Sbjct: 1288GKSPSKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGIFTRLISR 1458
Query: 425 NTPIPIKMTEDRCKTSVDNQL--GVSIHVYEGERIKASENNLLGLFRLF-VPPAPRGL-C 480
NT IP K ++ T+ DNQ G EG R A++N LG F L +PPAPRGL
Sbjct: 1459NTTIPTKKSQ-VFSTAADNQTQRGYXRCSQEGXREMAADNKSLGEFDLVGIPPAPRGLPQ 1635
Query: 481 IKVCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEAEKFKVEDMKFQ 540
I+V F IDA+GI+ V+A++++TG + +ITI + LS EI M++EAE D + +
Sbjct: 1636IEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSG-GLSDDEINNMVKEAELHAQRDQERK 1812
Query: 541 EKARAVNALEDYLYKTSKVMED 562
N+ + +Y K + +
Sbjct: 1813ALIDIKNSADTSIYSIEKSLSE 1878
>TC87268 homologue to PIR|S53500|S44168 dnaK-type molecular chaperone
HSC71.0 - garden pea, partial (59%)
Length = 1336
Score = 369 bits (948), Expect = e-102
Identities = 202/359 (56%), Positives = 259/359 (71%), Gaps = 3/359 (0%)
Frame = +2
Query: 267 KALRKLRTACERAKRALSYDIEATIDIDAICQGIDFCSSITRAKFERLNMDLFEKCMEIV 326
+ALR+LRTACERAKR LS + TI+ID++ +GIDF S ITRA+FE LNMDLF KCME V
Sbjct: 2 RALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYSPITRARFEELNMDLFRKCMEPV 181
Query: 327 RSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQDFFKGRDLCMSINPDEAVAYGAAVQA 386
CLRDA MD+ VH++VLVGGS+RIPKV++LLQDFF G++LC SINPDEAVAYGAAVQA
Sbjct: 182 EKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQA 361
Query: 387 ALLI-DGIKNVPNLVLRDITPLSLGIWTKGDVMSVVIPKNTPIPIKMTEDRCKTSVDNQL 445
A+L +G + V +L+L D+TPLSLG+ T G VM+V+IP+NT IP K E T DNQ
Sbjct: 362 AILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTK-KEQVFSTYSDNQP 538
Query: 446 GVSIHVYEGERIKASENNLLGLFRLF-VPPAPRGL-CIKVCFAIDADGILNVTAEEETTG 503
GV I V+EGER + +NNLLG F L +PPAPRG+ I VCF IDA+GILNV+AE++TTG
Sbjct: 539 GVLIQVFEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTG 718
Query: 504 NKSEITITNDNRRLSTQEIERMIQEAEKFKVEDMKFQEKARAVNALEDYLYKTSKVMEDN 563
K++ITITND RLS ++IE+M+QEAEK+K ED + ++K A NALE+Y Y ++D
Sbjct: 719 QKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDE 898
Query: 564 SVSSMLAPADKMKIDSAIMNGKSLIDNHGNQHNETYVFVDFLKELEIIFESSLNKINKG 622
++ L DK KI+ I +D NQ E F D +KELE + + K+ +G
Sbjct: 899 KIAGKLDSDDKKKIEDTIEAAIQWLD--ANQLAEADEFEDKMKELEGVCNPIIAKMYQG 1069
>TC80756 homologue to PIR|S53498|S53498 dnaK-type molecular chaperone
HSP71.2 - garden pea, partial (53%)
Length = 1189
Score = 320 bits (819), Expect = 1e-87
Identities = 175/320 (54%), Positives = 229/320 (70%), Gaps = 3/320 (0%)
Frame = +3
Query: 306 ITRAKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQDFFKG 365
ITRA+FE LNMDLF KCME V CLRDA +D+S VHE+VLVGGS+RIPKV++LLQDFF G
Sbjct: 3 ITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSHVHEVVLVGGSTRIPKVQQLLQDFFNG 182
Query: 366 RDLCMSINPDEAVAYGAAVQAALLI-DGIKNVPNLVLRDITPLSLGIWTKGDVMSVVIPK 424
++LC SINPDEAVAYGAAVQAA+L +G + V +L+L D+TPLSLG+ T G VM+ +IP+
Sbjct: 183 KELCKSINPDEAVAYGAAVQAAILSGEGDEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 362
Query: 425 NTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERIKASENNLLGLFRLF-VPPAPRGL-CIK 482
NT IP K E T DNQ GV I V+EGER + +NNLLG F L +PPAPRG+ I
Sbjct: 363 NTTIPTK-KEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQIN 539
Query: 483 VCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEAEKFKVEDMKFQEK 542
VCF IDA+GILNV+AE++T G K++ITITND RLS +EIE+M+++AEK+K ED + ++K
Sbjct: 540 VCFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAEKYKAEDEEVKKK 719
Query: 543 ARAVNALEDYLYKTSKVMEDNSVSSMLAPADKMKIDSAIMNGKSLIDNHGNQHNETYVFV 602
A N++E+Y Y ++D + L+ DK KI+ A+ + ++ GNQ E F
Sbjct: 720 VEAKNSIENYAYNMRNTIKDEKIGGKLSHEDKEKIEKAVEDAIQWLE--GNQMAEVDEFE 893
Query: 603 DFLKELEIIFESSLNKINKG 622
D KELE I + K+ +G
Sbjct: 894 DKQKELEGICNPIIAKMYQG 953
>BQ139103 homologue to GP|2655420|gb| heat shock cognate protein HSC70
{Brassica napus}, partial (29%)
Length = 628
Score = 283 bits (724), Expect = 1e-76
Identities = 142/193 (73%), Positives = 164/193 (84%)
Frame = +2
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAA 60
MA K EGPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS VAFTDS RLIG+AA
Sbjct: 56 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 229
Query: 61 KNQAASNPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCF 120
KNQ A NP NT+FDAKRLIGR+FSD+ ++ D+KLWPFK+I G + P I V YKG++K F
Sbjct: 230 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKIISGPAEKPLIGVNYKGEDKEF 409
Query: 121 VAEEISSMILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIIN 180
AEEISSM+L KM+EIAE +L +KNAV+TVPAYFNDSQR+ATKDAG+IAGLNVMRIIN
Sbjct: 410 AAEEISSMVLMKMREIAEAYLGSAIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 589
Query: 181 EPTAAALAYGIQK 193
EPTAAA+AYG+ K
Sbjct: 590 EPTAAAIAYGLDK 628
>TC85394 homologue to GP|2642238|gb|AAB86942.1| endoplasmic reticulum
HSC70-cognate binding protein precursor {Glycine max},
partial (38%)
Length = 962
Score = 277 bits (709), Expect = 8e-75
Identities = 146/231 (63%), Positives = 177/231 (76%), Gaps = 1/231 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAAKNQAAS 66
G IGIDLGTTYSCV V+ +N EII NDQGNR TPS V+FTD RLIG AAKN AA
Sbjct: 289 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 462
Query: 67 NPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYK-GKEKCFVAEEI 125
NP TIFD KRLIGRKF D +++D+KL P+K++ D P I V+ K G+ K F EEI
Sbjct: 463 NPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NRDGKPYIQVRVKDGETKVFSPEEI 639
Query: 126 SSMILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIINEPTAA 185
S+MIL KM+E AE FL +++AV+TVPAYFND+QR+ATKDAG+IAGLNV RIINEPTAA
Sbjct: 640 SAMILGKMKETAEGFLGKTIRDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 819
Query: 186 ALAYGIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTH 236
A+AYG+ K+G G++N+ +FDLGGGTFDVS+LT+ + FEV AT GDTH
Sbjct: 820 AIAYGLDKKG----GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTH 960
>TC77585 similar to GP|17473863|gb|AAL38353.1 putative heat-shock protein
{Arabidopsis thaliana}, partial (92%)
Length = 3027
Score = 252 bits (644), Expect = 3e-67
Identities = 150/432 (34%), Positives = 229/432 (52%), Gaps = 12/432 (2%)
Frame = +1
Query: 10 IGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAAKNQAASNPT 69
+G D G VAV +QR +++ ND+ R TP++V F D R IG A NP
Sbjct: 247 VGFDFGNESCIVAVARQRG--IDVVLNDESKRETPAIVCFGDKQRFIGTAGAASTMMNPK 420
Query: 70 NTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCFVAEEISSMI 129
N+I KRLIG+KF+D +++D+K PF V G D P I +Y G+ + F A ++ M+
Sbjct: 421 NSISQIKRLIGKKFADPELQRDLKSLPFNVTEGPDGYPLIHARYLGESREFTATQVFGMM 600
Query: 130 LSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIINEPTAAALAY 189
LS ++EIA+K L V + I +P YF D QR++ DA IAGL+ + +I+E TA ALAY
Sbjct: 601 LSNLKEIAQKNLNAAVVDCCIGIPVYFTDLQRRSVLDAATIAGLHPLHLIHETTATALAY 780
Query: 190 GIQKRGNFVVGKRNVFIFDLGGGTFDVSLLTLKDDSFEVKATAGDTHLGGEDFDNKMVSH 249
GI K NV D+G + V + K V + + D LGG DFD + H
Sbjct: 781 GIYKTDLPENEWLNVAFVDVGHASMQVCIAGFKKGQLHVLSHSYDRSLGGRDFDEALFHH 960
Query: 250 FVNELKRKNNVDISLNPKALRKLRTACERAKRALSYDIEATIDIDAICQGIDFCSSITRA 309
F + K + +D+ N +A +LR ACE+ K+ LS + EA ++I+ + D I R
Sbjct: 961 FAAKFKEEYKIDVYQNARACLRLRAACEKLKKVLSANPEAPLNIECLMDEKDVRGFIKRD 1140
Query: 310 KFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKLLQDFFKGRDLC 369
FE+L++ + E+ + L +A + +H + +VG SR+P + K+L +FFK ++
Sbjct: 1141DFEQLSLPILERVKGPLEKALAEAGLTVENIHMVEVVGSGSRVPAINKILTEFFK-KEPR 1317
Query: 370 MSINPDEAVAYGAAVQAALLIDGIKNVPNLVLRDITPLSLGI-W-----------TKGDV 417
++N E VA GAA+Q A+L K V + + P S+ + W +
Sbjct: 1318RTMNASECVARGAALQCAILSPTFK-VREFQVNESFPFSVSLSWKYSGSDAPDSESDNKQ 1494
Query: 418 MSVVIPKNTPIP 429
++V PK PIP
Sbjct: 1495STIVFPKGNPIP 1530
>TC80052 similar to PIR|JC4786|JC4786 dnaK-type molecular chaperone hsc70-3
- tomato, partial (27%)
Length = 667
Score = 250 bits (638), Expect = 1e-66
Identities = 131/186 (70%), Positives = 150/186 (80%)
Frame = +3
Query: 9 AIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAAKNQAASNP 68
AIGIDLGTTYSCVAV +N +II ND G RTTPS VAF DS R+IG+AA N AASNP
Sbjct: 105 AIGIDLGTTYSCVAVC--KNGEIDIIVNDLGKRTTPSFVAFKDSERMIGDAAFNIAASNP 278
Query: 69 TNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCFVAEEISSM 128
TNTIFDAKRLIGRKFSD I++ D+KLWPFKVI +D P I+V Y +EK F AEEISSM
Sbjct: 279 TNTIFDAKRLIGRKFSDPIVQSDVKLWPFKVIGDLNDKPMIVVNYNDEEKHFAAEEISSM 458
Query: 129 ILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDAGIIAGLNVMRIINEPTAAALA 188
+L KM+EIAE FL V++ VITVPAYFNDSQR++T+DAG IAGLNVMRIINEPTAAA+A
Sbjct: 459 VLVKMREIAETFLGSIVEDVVITVPAYFNDSQRQSTRDAGAIAGLNVMRIINEPTAAAIA 638
Query: 189 YGIQKR 194
YG +
Sbjct: 639 YGFNTK 656
>TC85391 homologue to PIR|T06598|T06598 dnaK-type molecular chaperone BiP-A
- soybean, partial (52%)
Length = 1231
Score = 249 bits (635), Expect = 3e-66
Identities = 145/317 (45%), Positives = 209/317 (65%), Gaps = 4/317 (1%)
Frame = +1
Query: 299 GIDFCSSITRAKFERLNMDLFEKCMEIVRSCLRDAYMDQSMVHEIVLVGGSSRIPKVRKL 358
G+ F +TRA+FE LN DLF K M V+ + DA + ++ + EIVLVGGS+RIPKV++L
Sbjct: 4 GVAFSEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKNQIDEIVLVGGSTRIPKVQQL 183
Query: 359 LQDFFKGRDLCMSINPDEAVAYGAAVQAALLI-DGIKNVPNLVLRDITPLSLGIWTKGDV 417
L+D+F G++ +NPDEAVA+GAAVQ ++L +G +++L D+ PL+LGI T G V
Sbjct: 184 LKDYFDGKEPNKGVNPDEAVAFGAAVQGSILSGEGGAETKDILLLDVAPLTLGIETVGGV 363
Query: 418 MSVVIPKNTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERIKASENNLLGLFRLF-VPPAP 476
M+ +IP+NT IP K ++ T D Q VSI V+EGER + LLG F L +PPAP
Sbjct: 364 MTKLIPRNTVIPTKKSQ-VFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAP 540
Query: 477 RGL-CIKVCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEAEKFKVE 535
RG I+V F +DA+GILNV AE++ TG +ITITN+ RLS +EIERM++EAE+F E
Sbjct: 541 RGTPQIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEE 720
Query: 536 DMKFQEKARAVNALEDYLYK-TSKVMEDNSVSSMLAPADKMKIDSAIMNGKSLIDNHGNQ 594
D K +E+ A NALE Y+Y ++V + + ++ L +K KI++A+ +D+ NQ
Sbjct: 721 DKKVKERIDARNALETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDD--NQ 894
Query: 595 HNETYVFVDFLKELEII 611
E + + LKE+E +
Sbjct: 895 SVEKEEYEEKLKEVEAV 945
>AW559598 weakly similar to SP|P09189|HS7C Heat shock cognate 70 kDa protein.
[Petunia] {Petunia hybrida}, partial (24%)
Length = 532
Score = 234 bits (597), Expect = 7e-62
Identities = 114/167 (68%), Positives = 136/167 (81%)
Frame = +1
Query: 1 MAKKYEGPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAA 60
MA E A+GIDLGTTYSCVAVW + NR EIIHND+G++ TPS VAFTD RL+G AA
Sbjct: 31 MANNCECYAVGIDLGTTYSCVAVWLEEKNRVEIIHNDEGSKITPSFVAFTDGQRLVGAAA 210
Query: 61 KNQAASNPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCF 120
K+QAA NP NT+FDAKRLIGRKFSD +++KDI LWPFKV+ G +D P I +K+KG+EK
Sbjct: 211 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDILLWPFKVVSGVNDKPMISLKFKGQEKLL 390
Query: 121 VAEEISSMILSKMQEIAEKFLELPVKNAVITVPAYFNDSQRKATKDA 167
AEEISS++L+ M+EIAE +LE PVKNA ITVPAYFND+QRKAT DA
Sbjct: 391 CAEEISSIVLTNMREIAEMYLESPVKNAGITVPAYFNDAQRKATIDA 531
>TC85395 homologue to GP|6911553|emb|CAB72130.1 heat shock protein 70
{Cucumis sativus}, partial (35%)
Length = 723
Score = 205 bits (522), Expect = 4e-53
Identities = 117/227 (51%), Positives = 154/227 (67%), Gaps = 2/227 (0%)
Frame = +3
Query: 398 NLVLRDITPLSLGIWTKGDVMSVVIPKNTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERI 457
+L+L D+TPLSLG+ T G VM+V+IP+NT IP K E T DNQ GV I VYEGER
Sbjct: 6 DLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKK-EQVFSTYSDNQPGVLIQVYEGERA 182
Query: 458 KASENNLLGLFRLF-VPPAPRGLC-IKVCFAIDADGILNVTAEEETTGNKSEITITNDNR 515
+ +NNLLG F L +PPAPRG+ I VCF IDA+GILNV+AE++TTG K++ITITND
Sbjct: 183 RTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKG 362
Query: 516 RLSTQEIERMIQEAEKFKVEDMKFQEKARAVNALEDYLYKTSKVMEDNSVSSMLAPADKM 575
RLS +EIE+M++EAEK+K ED + ++K A NALE+Y Y ++D + S L+P DK
Sbjct: 363 RLSKEEIEKMVEEAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIGSKLSPEDKK 542
Query: 576 KIDSAIMNGKSLIDNHGNQHNETYVFVDFLKELEIIFESSLNKINKG 622
KID AI +D+ NQ E F D +KELE + + K+ +G
Sbjct: 543 KIDDAIDAAIQWLDS--NQLAEADEFQDKMKELESLCNPIIAKMYQG 677
>BQ752418 homologue to SP|P78695|GR78 78 kDa glucose-regulated protein
homolog precursor (GRP 78), partial (47%)
Length = 1032
Score = 187 bits (476), Expect(3) = 2e-51
Identities = 107/248 (43%), Positives = 157/248 (63%), Gaps = 3/248 (1%)
Frame = -1
Query: 352 IPKVRKLLQDFFKGRDLCMSINPDEAVAYGAAVQAALLIDGIKNVPNLVLRDITPLSLGI 411
IPKV+ L++++F G+ INPDEAVA+GAAVQA +L G + +VL D+ PL+LGI
Sbjct: 900 IPKVQSLIEEYFGGKKASKGINPDEAVAFGAAVQAGVL-SGEEGTEEIVLMDVNPLTLGI 724
Query: 412 WTKGDVMSVVIPKNTPIPIKMTEDRCKTSVDNQLGVSIHVYEGERIKASENNLLGLFRLF 471
T G VM+ +I +NTPIP + ++ T+ DNQ V I V+EGER +NN LG F L
Sbjct: 723 ETTGGVMTKLITRNTPIPTRKSQI-FSTAADNQPVVLIQVFEGERSLTKDNNQLGKFELT 547
Query: 472 -VPPAPRGLC-IKVCFAIDADGILNVTAEEETTGNKSEITITNDNRRLSTQEIERMIQEA 529
+PPAPRG+ I+V F +DA+GIL V+A ++ TG + ITITND RL+ +EI+RM++EA
Sbjct: 546 GIPPAPRGVPQIEVSFELDANGILKVSAHDKGTGKQESITITNDKGRLTKEEIDRMVEEA 367
Query: 530 EKFKVEDMKFQEKARAVNALEDYLYK-TSKVMEDNSVSSMLAPADKMKIDSAIMNGKSLI 588
EK+ ED +E+ A N LE+Y + ++V ++ + + DK I A+ +
Sbjct: 366 EKYAEEDKATRERIEARNGLENYAFSLKNQVNDEEGLGGKIDDEDKETILEAVKETNDWL 187
Query: 589 DNHGNQHN 596
+ +G N
Sbjct: 186 EENGATAN 163
Score = 29.6 bits (65), Expect(3) = 2e-51
Identities = 14/29 (48%), Positives = 20/29 (68%)
Frame = -3
Query: 326 VRSCLRDAYMDQSMVHEIVLVGGSSRIPK 354
V L+DA + + V +IVLVGGS+R P+
Sbjct: 979 VEQVLKDAKVKKEDVDDIVLVGGSTRYPQ 893
Score = 25.0 bits (53), Expect(3) = 2e-51
Identities = 10/16 (62%), Positives = 13/16 (80%)
Frame = -2
Query: 309 AKFERLNMDLFEKCME 324
AKFE LNMDL +K ++
Sbjct: 1031 AKFEELNMDLLKKTLK 984
>CA859549 similar to GP|20260807|gb Hsp70 protein 1 {Rhizopus stolonifer},
partial (19%)
Length = 527
Score = 169 bits (427), Expect = 4e-42
Identities = 87/127 (68%), Positives = 101/127 (79%)
Frame = +3
Query: 6 EGPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAAKNQAA 65
EG A+GIDLGTTYSCV VWQ N+R EII NDQGNRTTPS VAFTD+ RLIG+AAKNQ A
Sbjct: 156 EGKAVGIDLGTTYSCVGVWQ--NDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVA 329
Query: 66 SNPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYKGKEKCFVAEEI 125
NP NT+FDAKRLIGR+F+D ++ D+K WPFKVI A P I V+YKG+ K F EEI
Sbjct: 330 MNPYNTVFDAKRLIGRRFADQEVQSDMKHWPFKVIDKA-AKPYIQVEYKGETKQFTPEEI 506
Query: 126 SSMILSK 132
SSM+L+K
Sbjct: 507 SSMVLTK 527
>AW695262 homologue to GP|170386|gb|A glucose-regulated protein 78
{Lycopersicon esculentum}, partial (39%)
Length = 607
Score = 167 bits (423), Expect = 1e-41
Identities = 92/158 (58%), Positives = 115/158 (72%), Gaps = 2/158 (1%)
Frame = +2
Query: 7 GPAIGIDLGTTYSCVAVWQQRNNRAEIIHNDQGNRTTPSVVAFTDSLRLIGNAAKNQAAS 66
G IGIDLGTTYSCV V+ +N EII NDQGNR TPS V+FTD RLIG AAKN AA
Sbjct: 128 GTVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVSFTDDERLIGEAAKNLAAV 301
Query: 67 NPTNTIFDAKRLIGRKFSDSIIRKDIKLWPFKVIPGADDNPKILVKYK-GKEKCFVAEEI 125
NP TIFD KRLIGRKF+D +++D+KL P+K++ D P I V+ K G+ K F EE+
Sbjct: 302 NPERTIFDVKRLIGRKFADKEVQRDMKLVPYKIV-NKDGKPYIQVRVKDGETKVFSPEEV 478
Query: 126 SSMILSKMQEIAEKFLELPVKNAVITVPAYF-NDSQRK 162
S+MIL+KM+E AE FL +++AV+TVPA ND+QR+
Sbjct: 479 SAMILTKMKETAEAFLGKTIRDAVVTVPALLSNDAQRQ 592
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,319,439
Number of Sequences: 36976
Number of extensions: 201583
Number of successful extensions: 924
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 831
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 834
length of query: 629
length of database: 9,014,727
effective HSP length: 102
effective length of query: 527
effective length of database: 5,243,175
effective search space: 2763153225
effective search space used: 2763153225
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149204.3


