

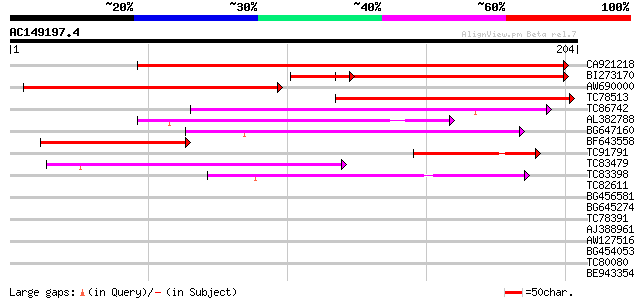
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.4 - phase: 0
(204 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA921218 180 4e-46
BI273170 104 2e-26
AW690000 91 3e-19
TC78513 homologue to GP|5679340|emb|CAB51773.1 hypothetical prot... 88 3e-18
TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG00... 79 9e-16
AL382788 weakly similar to GP|11322386|emb glycine receptor beta... 64 3e-11
BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrio... 58 2e-09
BF643558 58 2e-09
TC91791 weakly similar to GP|7769858|gb|AAF69536.1| F12M16.20 {A... 48 2e-06
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 44 6e-05
TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.... 42 2e-04
TC82611 32 0.001
BG456581 35 0.020
BG645274 32 0.13
TC78391 similar to PIR|F84919|F84919 glutathione-conjugate trans... 32 0.17
AJ388961 similar to GP|1492089|gb|A MC146R {Molluscum contagiosu... 32 0.22
AW127516 31 0.29
BG454053 similar to GP|19698833|gb 60S ribosomal protein L10A {A... 31 0.37
TC80080 similar to GP|9802755|gb|AAF99824.1| Unknown protein {Ar... 30 0.64
BE943354 30 0.83
>CA921218
Length = 707
Score = 180 bits (456), Expect = 4e-46
Identities = 81/155 (52%), Positives = 107/155 (68%)
Frame = -3
Query: 47 WIKGNCVGAFASGKAACGGIFRNRLGQFLGCFAEGLSYGNSLFAELCGAMRSIEFAQSKS 106
WI N G+ +ACGG FRN FL CFAE GN+L A+L GAMR+IE A +++
Sbjct: 582 WINCNTNGSANINTSACGGTFRNSNAYFLLCFAENTGNGNALHAKLSGAMRAIEIAAARN 403
Query: 107 WNNFWLETDSMLVVQAFKNSAIVPWQVRNRWNNCQIIISNMNFLASHIYREGNSCADVLA 166
W++ WLE DS LVV AFK+ +++ W +RNRWNNC +IS+MNF SH++REGN CAD LA
Sbjct: 402 WSHLWLELDSSLVVNAFKSKSVIHWNLRNRWNNCLFLISSMNFFVSHVFREGNQCADGLA 223
Query: 167 NIGLTLDTYVYYYSLPLQVRSDYVKNRLGWPSFRF 201
N GL+LD Y+ +P + YV+N++GWP FRF
Sbjct: 222 NFGLSLDHLTYWNHVPPFIHRFYVENKIGWPMFRF 118
>BI273170
Length = 391
Score = 104 bits (260), Expect(2) = 2e-26
Identities = 44/84 (52%), Positives = 62/84 (73%)
Frame = +1
Query: 118 LVVQAFKNSAIVPWQVRNRWNNCQIIISNMNFLASHIYREGNSCADVLANIGLTLDTYVY 177
L+ FKN + VPW++RNRW NC + NMNF+ SH++REGN CAD+LANIGL+L+
Sbjct: 52 LLSMPFKNISQVPWKLRNRWENCILATRNMNFIVSHVFREGNECADMLANIGLSLNCLTI 231
Query: 178 YYSLPLQVRSDYVKNRLGWPSFRF 201
+ LP ++S ++KN+LGWP+ RF
Sbjct: 232 WLELPDCIKSIFIKNKLGWPNNRF 303
Score = 30.8 bits (68), Expect(2) = 2e-26
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +3
Query: 102 AQSKSWNNFWLETDSMLVVQAFK 124
A+ +W + WLE+DS LVV AF+
Sbjct: 3 AKLHNWQSLWLESDSALVVNAFQ 71
>AW690000
Length = 652
Score = 90.9 bits (224), Expect = 3e-19
Identities = 46/93 (49%), Positives = 59/93 (62%)
Frame = +3
Query: 6 SKLSASSSITEFTILKAFNVSIHPPRSMLVKEVLWSPPIHSWIKGNCVGAFASGKAACGG 65
S AS+SI+ F ILK FNV+IHPP++ + EV+W PPI WIK N G+ S +ACGG
Sbjct: 309 SNAVASASISNFLILKKFNVTIHPPKAPKIIEVIWRPPIPHWIKCNTDGSSRSHSSACGG 488
Query: 66 IFRNRLGQFLGCFAEGLSYGNSLFAELCGAMRS 98
IFRN L CFAE N+ AEL +++S
Sbjct: 489 IFRNHDTDLLLCFAENTGECNAFHAELLXSLKS 587
>TC78513 homologue to GP|5679340|emb|CAB51773.1 hypothetical protein {Galega
orientalis}, partial (20%)
Length = 765
Score = 87.8 bits (216), Expect = 3e-18
Identities = 38/86 (44%), Positives = 58/86 (67%)
Frame = +2
Query: 118 LVVQAFKNSAIVPWQVRNRWNNCQIIISNMNFLASHIYREGNSCADVLANIGLTLDTYVY 177
LVV K++ +PW + NRWNN ++I+ + + SHIYREGN AD LAN GL+LD+ +
Sbjct: 404 LVVLPSKSTNQIPWNL*NRWNNVKVILQGLKCIISHIYREGNQVADTLANHGLSLDSIHF 583
Query: 178 YYSLPLQVRSDYVKNRLGWPSFRFNH 203
+ +P R+ Y +N+ GW +FRF++
Sbjct: 584 WNDVPEYTRASYFRNKDGWSNFRFSY 661
>TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG005I10.16 -
Arabidopsis thaliana, partial (19%)
Length = 2073
Score = 79.3 bits (194), Expect = 9e-16
Identities = 48/132 (36%), Positives = 69/132 (51%), Gaps = 2/132 (1%)
Frame = -1
Query: 66 IFRNRLGQFLGCFAEGLSYGNSLFAELCGAMRSIEFAQSKSWNNFWLETDSMLVVQAFKN 125
+ R+ FLG + + + L AE C M +IE A NN LETDS+ VV AF
Sbjct: 468 VIRDSQFGFLGALSCNIGHATPLEAEFCACMIAIEKAMELGLNNICLETDSLKVVNAFHK 289
Query: 126 SAIVPWQVRNRWNNCQIIISNMNFLASHIYREGNSCADVLA--NIGLTLDTYVYYYSLPL 183
+PWQ+R RW+NC ++ + HI REGN AD LA GL+L ++ + P
Sbjct: 288 IVGIPWQMRVRWHNCIRFCHSIACVCVHIPREGNLVADALARHGQGLSLFFLQWWPAPPS 109
Query: 184 QVRSDYVKNRLG 195
++S ++R G
Sbjct: 108 FIQSFLAQDRYG 73
>AL382788 weakly similar to GP|11322386|emb glycine receptor betaZ subunit
{Danio rerio}, partial (4%)
Length = 353
Score = 64.3 bits (155), Expect = 3e-11
Identities = 39/116 (33%), Positives = 56/116 (47%), Gaps = 2/116 (1%)
Frame = +2
Query: 47 WIKGNCVGAF--ASGKAACGGIFRNRLGQFLGCFAEGLSYGNSLFAELCGAMRSIEFAQS 104
WIK A +S + G +FR++ G GCF ++ AE A ++E
Sbjct: 11 WIKAKIDNATCGSSDPSIVGSLFRDQFGSNFGCFIIFFGVNFAIHAEFYAAAYAVEIVYL 190
Query: 105 KSWNNFWLETDSMLVVQAFKNSAIVPWQVRNRWNNCQIIISNMNFLASHIYREGNS 160
+W+NFWL D LVV AF ++ V W + + + MNF A HIY+E NS
Sbjct: 191 CNWHNFWLACDLKLVVDAFHSNYKVLWSLAT*F-----FCNQMNF*AFHIYKERNS 343
>BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrioides},
partial (2%)
Length = 780
Score = 58.2 bits (139), Expect = 2e-09
Identities = 38/123 (30%), Positives = 60/123 (47%), Gaps = 1/123 (0%)
Frame = +1
Query: 64 GGIFRNRLGQFLGCFAEGLS-YGNSLFAELCGAMRSIEFAQSKSWNNFWLETDSMLVVQA 122
GG+ RN L ++ F+ +S Y N +FAEL +S++ A S + +DS+L V
Sbjct: 379 GGVIRNYLSTYITGFSGFISIYQNIMFAELTTLHQSLKLAISLNIEEMVCYSDSLLTVNL 558
Query: 123 FKNSAIVPWQVRNRWNNCQIIISNMNFLASHIYREGNSCADVLANIGLTLDTYVYYYSLP 182
K +N + I+S+ NF H REGN CAD + + + D + +S P
Sbjct: 559 IKEDISQHHVYAVLIHNIKYIMSSRNFTLHHTLREGNQCADFMVKLRTSTDVDLTIHSSP 738
Query: 183 LQV 185
+V
Sbjct: 739 PKV 747
>BF643558
Length = 645
Score = 58.2 bits (139), Expect = 2e-09
Identities = 24/54 (44%), Positives = 37/54 (68%)
Frame = +2
Query: 12 SSITEFTILKAFNVSIHPPRSMLVKEVLWSPPIHSWIKGNCVGAFASGKAACGG 65
++IT+F LK F++ IHPP++ + EVLW+PP WIK N G+ + ++CGG
Sbjct: 473 TNITDFVFLKKFSIDIHPPKAPKIIEVLWNPPPLHWIKCNTDGSSNTNTSSCGG 634
>TC91791 weakly similar to GP|7769858|gb|AAF69536.1| F12M16.20 {Arabidopsis
thaliana}, partial (4%)
Length = 576
Score = 48.1 bits (113), Expect = 2e-06
Identities = 23/46 (50%), Positives = 32/46 (69%)
Frame = -2
Query: 146 NMNFLASHIYREGNSCADVLANIGLTLDTYVYYYSLPLQVRSDYVK 191
NMN + +HIY+EGN AD LAN GLTL++YV + + V+ Y+K
Sbjct: 572 NMNCIVTHIYKEGNQVADSLANFGLTLNSYV--FKVISNVKQIYIK 441
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 1222
Score = 43.5 bits (101), Expect = 6e-05
Identities = 30/110 (27%), Positives = 46/110 (41%), Gaps = 2/110 (1%)
Frame = +2
Query: 14 ITEFTILKAFN--VSIHPPRSMLVKEVLWSPPIHSWIKGNCVGAFASGKAACGGIFRNRL 71
I +LK N +I P S + W P W K N G+ A GG+ R+
Sbjct: 893 IKGMVLLKNLNQIANILNPVSRSIIWCEWKKPEIGWTKLNTDGSVNKETAGFGGLLRDYR 1072
Query: 72 GQFLGCFAEGLSYGNSLFAELCGAMRSIEFAQSKSWNNFWLETDSMLVVQ 121
G+ + F G++ EL R + + + W+E+DSM VV+
Sbjct: 1073 GEPICAFVSKAPQGDTFLVELWAIWRGLVLSLGLGIKSIWVESDSMSVVK 1222
>TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.40 -
Arabidopsis thaliana, partial (6%)
Length = 766
Score = 42.0 bits (97), Expect = 2e-04
Identities = 31/117 (26%), Positives = 55/117 (46%), Gaps = 1/117 (0%)
Frame = +2
Query: 72 GQFLGCFAEGLSYGNS-LFAELCGAMRSIEFAQSKSWNNFWLETDSMLVVQAFKNSAIVP 130
G F F+ + + N LFAEL + ++ AQ+ + + +DS+ V ++V
Sbjct: 89 GFFNSGFSGHIDHSNDILFAELHAILMGLQLAQTLNIVDVVCYSDSLHYVNLINGPSVVY 268
Query: 131 WQVRNRWNNCQIIISNMNFLASHIYREGNSCADVLANIGLTLDTYVYYYSLPLQVRS 187
+ + +I H REGN CAD LA +G ++D+ + ++ PL + S
Sbjct: 269 HAYATLIQDIKDLIRLSKL---HTLREGNRCADFLAKLGASVDSTLPIHATPLWIAS 430
>TC82611
Length = 814
Score = 32.0 bits (71), Expect(2) = 0.001
Identities = 12/22 (54%), Positives = 15/22 (67%)
Frame = +2
Query: 136 RWNNCQIIISNMNFLASHIYRE 157
RWNNC + S M F SHI+R+
Sbjct: 530 RWNNCINLSSRMRFYCSHIFRD 595
Score = 26.6 bits (57), Expect(2) = 0.001
Identities = 12/22 (54%), Positives = 16/22 (72%)
Frame = +3
Query: 114 TDSMLVVQAFKNSAIVPWQVRN 135
+DS LVV AFKN IV W+ ++
Sbjct: 468 SDSSLVV*AFKNMDIVSWKFKD 533
>BG456581
Length = 683
Score = 35.0 bits (79), Expect = 0.020
Identities = 16/47 (34%), Positives = 21/47 (44%)
Frame = +2
Query: 19 ILKAFNVSIHPPRSMLVKEVLWSPPIHSWIKGNCVGAFASGKAACGG 65
IL F + H + V W PP W+KGN G+ + A GG
Sbjct: 539 ILDVFRIPPHRRSMXEMVSVCWKPPSAPWVKGNTDGSXLNNSGAXGG 679
>BG645274
Length = 641
Score = 32.3 bits (72), Expect = 0.13
Identities = 15/32 (46%), Positives = 20/32 (61%)
Frame = -1
Query: 12 SSITEFTILKAFNVSIHPPRSMLVKEVLWSPP 43
S++ EF IL+ F V H P + +K VLW PP
Sbjct: 461 SNM*EFEILRVFFV*DHAPNASRIKAVLWYPP 366
>TC78391 similar to PIR|F84919|F84919 glutathione-conjugate transporter AtMRP4
[imported] - Arabidopsis thaliana, partial (17%)
Length = 1265
Score = 32.0 bits (71), Expect = 0.17
Identities = 19/74 (25%), Positives = 32/74 (42%)
Frame = +3
Query: 129 VPWQVRNRWNNCQIIISNMNFLASHIYREGNSCADVLANIGLTLDTYVYYYSLPLQVRSD 188
+PWQ+ N W NC I N+N I+ EG L +V ++ + + +
Sbjct: 1053 MPWQIHNSWFNCNRISGNINTFV-QIFVEGGLVGVDWQEWSL----FV*FFPAMVVLNTS 1217
Query: 189 YVKNRLGWPSFRFN 202
++K W F F+
Sbjct: 1218 FLKTFPSWDRFDFS 1259
>AJ388961 similar to GP|1492089|gb|A MC146R {Molluscum contagiosum virus
subtype 1}, partial (4%)
Length = 325
Score = 31.6 bits (70), Expect = 0.22
Identities = 14/34 (41%), Positives = 19/34 (55%)
Frame = -2
Query: 129 VPWQVRNRWNNCQIIISNMNFLASHIYREGNSCA 162
+PWQ+ N W NC I N+N I+ EG+ A
Sbjct: 159 MPWQIHNSWFNCNRISGNINTFV-QIFIEGSLVA 61
>AW127516
Length = 379
Score = 31.2 bits (69), Expect = 0.29
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +3
Query: 144 ISNMNFLASHIYREGNSCADVLANIG 169
I++ +F H REGN CAD +A +G
Sbjct: 216 IASRSFTIQHTLREGNHCADYMAKLG 293
>BG454053 similar to GP|19698833|gb 60S ribosomal protein L10A {Arabidopsis
thaliana}, partial (96%)
Length = 669
Score = 30.8 bits (68), Expect = 0.37
Identities = 15/41 (36%), Positives = 24/41 (57%), Gaps = 1/41 (2%)
Frame = -3
Query: 17 FTILKAFNVSIHPPRSMLVKEVLWSPPIHSWIKGNCV-GAF 56
F+ L++ V + PR++ + LWSP H+ I G V G+F
Sbjct: 319 FSFLRSSTVMVSMPRALASSQCLWSPSTHTAILGRGVYGSF 197
>TC80080 similar to GP|9802755|gb|AAF99824.1| Unknown protein {Arabidopsis
thaliana}, partial (5%)
Length = 1149
Score = 30.0 bits (66), Expect = 0.64
Identities = 13/29 (44%), Positives = 20/29 (68%), Gaps = 1/29 (3%)
Frame = +1
Query: 129 VPWQVRNRWNNCQIIISNMN-FLASHIYR 156
+PW+++N W NC + SN+N FL + I R
Sbjct: 832 LPWKLQNSWFNCIRLASNINHFLPTFIER 918
>BE943354
Length = 361
Score = 29.6 bits (65), Expect = 0.83
Identities = 14/31 (45%), Positives = 16/31 (51%)
Frame = -3
Query: 128 IVPWQVRNRWNNCQIIISNMNFLASHIYREG 158
IVPW + NRW NC + NF REG
Sbjct: 356 IVPWDLCNRWRNC-FGLHKFNFYFFMFSREG 267
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.135 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,136,972
Number of Sequences: 36976
Number of extensions: 126428
Number of successful extensions: 664
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 659
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 663
length of query: 204
length of database: 9,014,727
effective HSP length: 92
effective length of query: 112
effective length of database: 5,612,935
effective search space: 628648720
effective search space used: 628648720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149197.4


