

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.10 - phase: 0
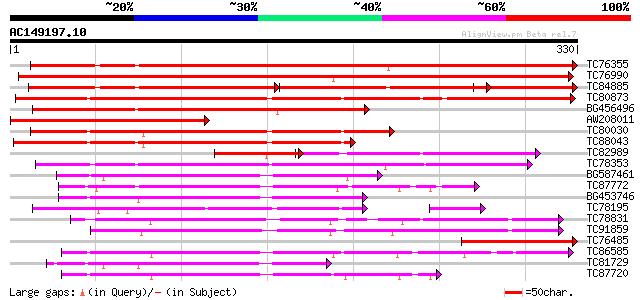
(330 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76355 similar to GP|16604487|gb|AAL24249.1 At2g45600/F17K2.13 ... 397 e-111
TC76990 similar to GP|16604487|gb|AAL24249.1 At2g45600/F17K2.13 ... 348 2e-96
TC84885 similar to GP|16604487|gb|AAL24249.1 At2g45600/F17K2.13 ... 173 3e-86
TC80873 weakly similar to GP|18086341|gb|AAL57633.1 AT3g48690/T8... 276 1e-74
BG456496 weakly similar to GP|10120426|gb| Hypothetical protein ... 206 8e-54
AW208011 similar to GP|16604487|gb| At2g45600/F17K2.13 {Arabidop... 195 2e-50
TC80030 weakly similar to PIR|T46214|T46214 hypothetical protein... 184 4e-47
TC88043 weakly similar to PIR|B96533|B96533 hypothetical protein... 174 6e-44
TC82989 weakly similar to PIR|A96515|A96515 hypothetical protein... 106 6e-36
TC78353 similar to GP|10178113|dbj|BAB11406. gb|AAD04946.2~gene_... 139 1e-33
BG587461 weakly similar to GP|8809631|dbj| HSR203J protein-like ... 124 7e-29
TC87772 weakly similar to GP|8574455|gb|AAF77578.1| pepper ester... 122 1e-28
BG453746 weakly similar to GP|8574455|gb|A pepper esterase {Caps... 117 6e-27
TC78195 similar to GP|6092014|dbj|BAA85654.1 hsr203J homolog {Pi... 106 4e-26
TC78831 similar to GP|8809707|dbj|BAA97248.1 contains similarity... 103 9e-23
TC91859 similar to GP|8809707|dbj|BAA97248.1 contains similarity... 101 4e-22
TC76485 homologue to GP|4097684|gb|AAD00154.1| ubiquitin conjuga... 100 1e-21
TC86585 weakly similar to GP|8809631|dbj|BAA97182.1 HSR203J prot... 99 3e-21
TC81729 weakly similar to GP|8574455|gb|AAF77578.1| pepper ester... 98 4e-21
TC87720 weakly similar to GP|8574455|gb|AAF77578.1| pepper ester... 96 2e-20
>TC76355 similar to GP|16604487|gb|AAL24249.1 At2g45600/F17K2.13
{Arabidopsis thaliana}, partial (17%)
Length = 1090
Score = 397 bits (1021), Expect = e-111
Identities = 187/320 (58%), Positives = 239/320 (74%), Gaps = 2/320 (0%)
Frame = +2
Query: 13 TKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLP 72
TK I++E+ + ++SDG+++RP+Q+P PP+ + PN+ SKDI I NP IS+RIYLP
Sbjct: 20 TKHIISEIPTYITVYSDGTVDRPRQAPTVPPNPDHPNS--PSKDIIISQNPNISARIYLP 193
Query: 73 KITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPT 132
K NP +K PILV+FHGG F FES FSK YHEH F QAN I+VS+EY LAPE+PLP
Sbjct: 194 K--NPTTKLPILVFFHGGGFFFESAFSKLYHEHFNVFVPQANSIVVSVEYRLAPEHPLPA 367
Query: 133 CYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENL 192
CY+DCW +L+W++S+S N NPE WLI HG+FN++FIGGDSAG NI HNIA++AG E L
Sbjct: 368 CYNDCWNSLQWVASNSAPNPVNPESWLINHGDFNRVFIGGDSAGGNIVHNIAMRAGSEAL 547
Query: 193 PCDVKILGAIIIHPYFYSANPIGSEPI--IEPENNIIHTFWHFAYPNAPFGIDNPRFNPL 250
P VK+LGAI+ PYFYS+ P+G E + + + ++ W+F YP+AP GIDNP NP+
Sbjct: 548 PNGVKLLGAILQQPYFYSSYPVGLESVKLKSSDKDFHYSVWNFVYPSAPGGIDNPMINPV 727
Query: 251 GEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLV 310
G GAPSL+ LGC RII+CVAGKD +RERGVWY+E VK SGWKGKLE FEE+DE HVY +
Sbjct: 728 GIGAPSLDGLGCDRIIICVAGKDGIRERGVWYYELVKKSGWKGKLELFEEEDEDHVYHIF 907
Query: 311 KPESESAKIFIQRLVGFVQE 330
PESES + I+ L F+ E
Sbjct: 908 HPESESGQKLIKHLASFLHE 967
>TC76990 similar to GP|16604487|gb|AAL24249.1 At2g45600/F17K2.13
{Arabidopsis thaliana}, partial (11%)
Length = 1264
Score = 348 bits (893), Expect = 2e-96
Identities = 164/325 (50%), Positives = 226/325 (69%), Gaps = 2/325 (0%)
Frame = +1
Query: 6 PTTPKTITKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTI 65
P++ + TKEI E+ +LR++ DG++ER S PP DP TG+SSKDI NP I
Sbjct: 43 PSSMASSTKEIDRELPPLLRVYKDGTVERFLGSKIVPPIPLDPETGVSSKDITFSQNPLI 222
Query: 66 SSRIYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLA 125
S+RI+LPK+TN K PILVY+HGG F ES FS + +L ASQANV++VS+EY LA
Sbjct: 223 SARIHLPKLTNQTQKLPILVYYHGGAFCLESAFSFLHQRYLNIIASQANVLVVSVEYRLA 402
Query: 126 PEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAI 185
PE+PLP Y D W +LKWI+SHS NNINN EPWLI++G+F++ +IGGD++GANIAHN +
Sbjct: 403 PEHPLPAAYDDGWFSLKWITSHSINNINNAEPWLIKYGDFDRFYIGGDTSGANIAHNALL 582
Query: 186 QA--GLENLPCDVKILGAIIIHPYFYSANPIGSEPIIEPENNIIHTFWHFAYPNAPFGID 243
+ G+E LP DVKI GA++ P F+S+ P+ SE + E + W+F YP+AP GID
Sbjct: 583 RVGNGVETLPGDVKIRGALLAFPLFWSSKPVLSESVEGHEQSSPMKVWNFVYPDAPGGID 762
Query: 244 NPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDE 303
NP NPL APSL+ +GC +I++ VAG D LR+RG+WY++ VK SGWKG +E + E
Sbjct: 763 NPLINPLAIDAPSLDIIGCPKILIFVAGNDDLRDRGIWYYDAVKKSGWKGDVELVHVEGE 942
Query: 304 GHVYQLVKPESESAKIFIQRLVGFV 328
H +Q+ PE++S+ ++R+ F+
Sbjct: 943 EHCFQIYHPETQSSIDMVKRIASFL 1017
>TC84885 similar to GP|16604487|gb|AAL24249.1 At2g45600/F17K2.13
{Arabidopsis thaliana}, partial (17%)
Length = 1102
Score = 173 bits (439), Expect(2) = 3e-86
Identities = 80/146 (54%), Positives = 106/146 (71%)
Frame = +2
Query: 12 ITKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYL 71
+ K I++E+ + ++SDG+++RP+Q P PP+ N PN+ SKDI I NP IS+RIYL
Sbjct: 92 VPKHIISEIPTYITVYSDGTVDRPRQPPTVPPNPNHPNS--PSKDIIISQNPNISARIYL 265
Query: 72 PKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLP 131
PK NP +K PILV+FHGG F FES FSK +HEH F AN I+VS+EY LAPE+PLP
Sbjct: 266 PK--NPTTKLPILVFFHGGGFFFESAFSKVHHEHFNIFIPLANSIVVSVEYRLAPEHPLP 439
Query: 132 TCYHDCWAALKWISSHSNNNINNPEP 157
CY+DCW +L+W++S+S N NPEP
Sbjct: 440 ACYNDCWNSLQWVASNSAKNPVNPEP 517
Score = 162 bits (411), Expect(2) = 3e-86
Identities = 78/123 (63%), Positives = 94/123 (76%)
Frame = +3
Query: 158 WLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIHPYFYSANPIGSE 217
WLI HG+FN++FIGG SAG NI HNIA++AG E LP DVK+LGAI+ HP FYS+ P+G E
Sbjct: 519 WLINHGDFNRVFIGGASAGGNIVHNIAMRAGSEALPNDVKLLGAILQHPLFYSSYPVGLE 698
Query: 218 PIIEPENNIIHTFWHFAYPNAPFGIDNPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRE 277
+ + W+F YP+AP GIDNP NP+G GAPSL+ LGC R+IVCVAGKDKLRE
Sbjct: 699 NV--KLKDFYSYLWNFVYPSAPGGIDNPMVNPVGIGAPSLDGLGCDRMIVCVAGKDKLRE 872
Query: 278 RGV 280
RGV
Sbjct: 873 RGV 881
Score = 73.9 bits (180), Expect = 8e-14
Identities = 33/60 (55%), Positives = 40/60 (66%)
Frame = +1
Query: 271 GKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPESESAKIFIQRLVGFVQE 330
GK L G WY+E +K SGWKGKLE FEE+DE HVY + PESES + I+ L F+ E
Sbjct: 853 GKINLGREGFWYYELIKKSGWKGKLELFEEEDEDHVYHIFHPESESGQKLIKHLASFLHE 1032
>TC80873 weakly similar to GP|18086341|gb|AAL57633.1 AT3g48690/T8P19_200
{Arabidopsis thaliana}, partial (49%)
Length = 1236
Score = 276 bits (705), Expect = 1e-74
Identities = 139/327 (42%), Positives = 202/327 (61%), Gaps = 1/327 (0%)
Frame = +2
Query: 4 TTPTTPKTITKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNP 63
+T + TI EI ++ ILR++ G +E F PPSL D T + SKD+ I
Sbjct: 26 STMDSTSTINDEIAIDIPPILRVYKSGRVENLIGEEFLPPSL-DQATNVESKDVVISEEH 202
Query: 64 TISSRIYLPKITNP-LSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEY 122
IS+R+++PK +P + K P+ VYFHGG F E+ FS YH +L + S ANVI VS+ Y
Sbjct: 203 NISARLFIPKTNHPPIQKLPVFVYFHGGGFCIETPFSPCYHNYLNSVTSLANVIGVSVHY 382
Query: 123 SLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHN 182
APEYP+P + D W ALKW++SH N + WL ++ +F K+F+GGDSAGANI+H
Sbjct: 383 RRAPEYPVPIAHEDSWLALKWVASHVGG--NGSDEWLNQYADFEKVFLGGDSAGANISHY 556
Query: 183 IAIQAGLENLPCDVKILGAIIIHPYFYSANPIGSEPIIEPENNIIHTFWHFAYPNAPFGI 242
+ I+ G ENL VK+ G++ IHPYF+ + IGSE + IH W F+ P G
Sbjct: 557 LGIRVGKENLD-GVKLEGSVYIHPYFWGVDLIGSESNMAEFVEKIHNLWRFSCPTTT-GS 730
Query: 243 DNPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKD 302
D+P NP + P L KLGC R+++CVAG+D L++RG +Y E ++ SGW G +E E +D
Sbjct: 731 DDPLINPAND--PDLGKLGCKRLLICVAGQDILKDRGWYYKELLEKSGWGGVVEVIETED 904
Query: 303 EGHVYQLVKPESESAKIFIQRLVGFVQ 329
E HV+ + KP ++A + + ++V F++
Sbjct: 905 ENHVFHMFKPTCDNAAVLLNQVVSFIK 985
>BG456496 weakly similar to GP|10120426|gb| Hypothetical protein {Arabidopsis
thaliana}, partial (13%)
Length = 674
Score = 206 bits (525), Expect = 8e-54
Identities = 99/200 (49%), Positives = 144/200 (71%), Gaps = 4/200 (2%)
Frame = +2
Query: 14 KEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHN-PTISSRIYLP 72
KEI E+ ++R++ DG+IER S PPSL DP TG+SSKDI I +N P++S+RI+LP
Sbjct: 29 KEIEKELLPLIRVYKDGTIERLMSSSIVPPSLQDPQTGVSSKDIVISNNNPSLSARIFLP 208
Query: 73 KITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPT 132
K ++ KFPIL+YFH G F ES FS H +L S++N+I VSI+Y L P++PLP
Sbjct: 209 K-SHHNHKFPILLYFHAGAFCVESPFSFFCHRYLNLLVSESNIIAVSIDYRLLPQHPLPA 385
Query: 133 CYHDCWAALKWISSHSNNNINN---PEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGL 189
Y D W +L+W++SH++N+ N+ E WL ++G+FNK++IGGD GAN+AHN+ ++AG
Sbjct: 386 AYEDGWTSLQWVASHTSNDPNSSIEKEQWLQDYGDFNKVYIGGDVNGANLAHNLXMRAGT 565
Query: 190 ENLPCDVKILGAIIIHPYFY 209
E LP ++KILGA++ P+F+
Sbjct: 566 ETLPNNLKILGALLCXPFFW 625
>AW208011 similar to GP|16604487|gb| At2g45600/F17K2.13 {Arabidopsis
thaliana}, partial (8%)
Length = 405
Score = 195 bits (496), Expect = 2e-50
Identities = 93/116 (80%), Positives = 101/116 (86%)
Frame = +1
Query: 1 MASTTPTTPKTITKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIP 60
MAS T TTP+T+ KEIVTE G +RIFSDGS+ERP QSP PPSLNDPNTG+SSKDI+IP
Sbjct: 58 MASKTQTTPETVAKEIVTEFGTFIRIFSDGSVERPSQSPLVPPSLNDPNTGLSSKDIEIP 237
Query: 61 HNPTISSRIYLPKITNPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVI 116
HNPTISSRIYL KITNP SKFPILVYFHGG F+FES FSK YH+HLK FASQANVI
Sbjct: 238 HNPTISSRIYLLKITNPPSKFPILVYFHGGAFIFESAFSKLYHDHLKIFASQANVI 405
>TC80030 weakly similar to PIR|T46214|T46214 hypothetical protein T8P19.210
- Arabidopsis thaliana, partial (31%)
Length = 827
Score = 184 bits (467), Expect = 4e-47
Identities = 91/215 (42%), Positives = 134/215 (62%), Gaps = 3/215 (1%)
Frame = +2
Query: 13 TKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLP 72
+ E+V ++ +++I+ DG IER S PPS DP T + SKDI I + IS+RI++P
Sbjct: 167 SNEVVLDLSPMIKIYKDGHIERLIGSDIVPPSF-DPTTNVESKDILISKDQNISARIFIP 343
Query: 73 KITN---PLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYP 129
K+ N P K P+LVYFHGG F E+ FS YH L T S+ANVI VS++Y APE+P
Sbjct: 344 KLNNDQFPNQKLPLLVYFHGGGFCVETPFSPPYHNFLNTIVSKANVIAVSVDYRRAPEHP 523
Query: 130 LPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGL 189
LP Y D W +LKW+ SH + N + W+ + +F K+F GDSAGANIA+++AI+ G
Sbjct: 524 LPIAYEDSWTSLKWVVSHLHG--NGSDEWINRYADFGKMFFAGDSAGANIANHMAIRVGT 697
Query: 190 ENLPCDVKILGAIIIHPYFYSANPIGSEPIIEPEN 224
+ L + + +++H +F+ +GSE + E+
Sbjct: 698 QGLQ-GINLERIVLVHTFFWGVERVGSEATEKSEH 799
>TC88043 weakly similar to PIR|B96533|B96533 hypothetical protein F14J22.12
[imported] - Arabidopsis thaliana, partial (33%)
Length = 745
Score = 174 bits (440), Expect = 6e-44
Identities = 89/201 (44%), Positives = 128/201 (63%), Gaps = 2/201 (0%)
Frame = +2
Query: 3 STTPTTPKTITKEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHN 62
ST +T TI E+ ++ +L+++ G ++R + PPSL DP T + SKD+ I
Sbjct: 158 STMDSTSSTIDDEVAVDLTPVLKLYKSGRVQRLAGTEVLPPSL-DPKTNVESKDVVISEE 334
Query: 63 PTISSRIYLPKITN--PLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSI 120
IS+R+++PK TN P K P+LVY HGG F E+ FS YH +L + S ANVI VS+
Sbjct: 335 HNISARLFIPK-TNYPPTQKLPLLVYIHGGAFCIETPFSPNYHNYLNSVTSLANVIGVSV 511
Query: 121 EYSLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIA 180
Y APE+P+PT + D W ALKW++SH N + WL ++ +F K+F+ GDSAGANIA
Sbjct: 512 HYRRAPEHPVPTGHEDSWLALKWVASHVGG--NGSDEWLNQYADFEKVFLRGDSAGANIA 685
Query: 181 HNIAIQAGLENLPCDVKILGA 201
H+++I+ G ENL VK+ G+
Sbjct: 686 HHLSIRVGKENLD-GVKLEGS 745
>TC82989 weakly similar to PIR|A96515|A96515 hypothetical protein F16N3.25
[imported] - Arabidopsis thaliana, partial (36%)
Length = 655
Score = 106 bits (265), Expect(2) = 6e-36
Identities = 58/143 (40%), Positives = 82/143 (56%)
Frame = +1
Query: 167 KLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIHPYFYSANPIGSEPIIEPENNI 226
K F+ GDSAGANI + IA++ N KILG I+++PYF+ PIG E + + +
Sbjct: 238 KFFLAGDSAGANIGNYIALKDHNFNF----KILGLIMVNPYFWGKEPIGEETSDDLKRRM 405
Query: 227 IHTFWHFAYPNAPFGIDNPRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGV 286
+ +W P+ G D+P NP E AP LE LG +++V V KD L ERG Y +
Sbjct: 406 VDRWWELVCPSDQ-GNDDPLINPFVEEAPRLEGLGVEKVLVTVCEKDILIERGKLYHNKL 582
Query: 287 KNSGWKGKLEFFEEKDEGHVYQL 309
NSGWKG E +E + + HV+ +
Sbjct: 583 VNSGWKGTAELYEIQGKDHVFHI 651
Score = 62.0 bits (149), Expect(2) = 6e-36
Identities = 27/54 (50%), Positives = 37/54 (68%), Gaps = 2/54 (3%)
Frame = +3
Query: 120 IEYSLAPEYPLPTCYHDCWAALKWISSHS--NNNINNPEPWLIEHGNFNKLFIG 171
+ Y LAPE+PLP Y D W A++WI+SH+ N N+ E WL E +FNK+F+G
Sbjct: 90 VNYRLAPEHPLPAAYDDSWEAVQWIASHAAENGEENDYESWLKEKVDFNKVFLG 251
>TC78353 similar to GP|10178113|dbj|BAB11406.
gb|AAD04946.2~gene_id:F15M7.10~similar to unknown
protein {Arabidopsis thaliana}, partial (62%)
Length = 1269
Score = 139 bits (351), Expect = 1e-33
Identities = 89/293 (30%), Positives = 148/293 (50%), Gaps = 4/293 (1%)
Frame = +2
Query: 16 IVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKIT 75
+V + L+++SDGSI R F P++ D ++ KD +S R+Y P
Sbjct: 107 VVEDCMGFLQLYSDGSIYRSNDFKFQIPTIED--NSVTFKDCLFDKKFNLSIRLYKPN-N 277
Query: 76 NPLSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYH 135
N K P++++ HGG F F S H + A+ ++++ +Y LAPE+ LP
Sbjct: 278 NKNKKLPVIMFLHGGGFCFGSRTWPHIHNCCMSLATGLQALVIAPDYRLAPEHRLPAAID 457
Query: 136 DCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCD 195
D A++W+ ++ EPWL +F ++FI GDS+G NIAH +A++ G + + +
Sbjct: 458 DAVEAVRWLQRQGLSH-GYGEPWLTGDVDFGRVFIMGDSSGGNIAHQLAVRFGSDRMAIE 634
Query: 196 -VKILGAIIIHPYFYSANPIGSE--PIIEPEN-NIIHTFWHFAYPNAPFGIDNPRFNPLG 251
V+I G +++ P+F SE P + N +++ FW + P D+P NP G
Sbjct: 635 PVRIRGYVLMAPFFGGEIRTESEEGPSEQTLNLDLLDRFWRLSMPVGETR-DHPLANPFG 811
Query: 252 EGAPSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEG 304
G+P+LE++ I+V V G + L++R Y +K G K FE + G
Sbjct: 812 HGSPNLEEMKLDPILVIVGGNELLKDRAANYATRLKGIGKDIKYIEFEGCEHG 970
>BG587461 weakly similar to GP|8809631|dbj| HSR203J protein-like protein
{Arabidopsis thaliana}, partial (22%)
Length = 700
Score = 124 bits (310), Expect = 7e-29
Identities = 76/198 (38%), Positives = 111/198 (55%), Gaps = 8/198 (4%)
Frame = +2
Query: 28 SDGSIERPKQSPFAPPSLNDPNTGIS--SKDIQIPHNPTISSRIYLPKIT-NPLSKFPIL 84
S+G+I R ++ P PS N PN IS +KDI I + S+RI+LP+ SK P++
Sbjct: 47 SNGTITRLREDPHISPSSN-PNLPISVLTKDILINPSHNTSARIFLPRTALEHASKLPLI 223
Query: 85 VYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWI 144
VYFHGG F+ S S H + A+ N I+VSI+Y L+PE+ LP Y D AL WI
Sbjct: 224 VYFHGGGFILFSAASDFLHNYCSNLANDVNSIVVSIDYRLSPEHRLPAAYDDAIEALHWI 403
Query: 145 SSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLE-NLPCD----VKIL 199
+ P+ WL + +++ +I G SAGANIA++ ++ +E NL + +KI
Sbjct: 404 KT-------QPDDWLRNYADYSNCYIMGSSAGANIAYHTCLRVAVETNLNHEYLKAIKIR 562
Query: 200 GAIIIHPYFYSANPIGSE 217
G I+ P+F N + SE
Sbjct: 563 GFILSQPFFGGTNRVASE 616
>TC87772 weakly similar to GP|8574455|gb|AAF77578.1| pepper esterase
{Capsicum annuum}, partial (34%)
Length = 1313
Score = 122 bits (307), Expect = 1e-28
Identities = 88/261 (33%), Positives = 134/261 (50%), Gaps = 16/261 (6%)
Frame = +1
Query: 29 DGSIERPKQSPFAPPSLNDPN--TGISSKDIQIPHNPTISSRIYLPKITNPLSK-FPILV 85
+G+I R + P +PPS DPN T SKD+ + + +RIYLP P SK P++V
Sbjct: 232 NGTITRLDKYPQSPPS-QDPNLPTPSLSKDLTLNPSKHTWARIYLPH--KPTSKKLPLIV 402
Query: 86 YFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWIS 145
++HGG F+F S S +H A+Q + ++VS+EY LAPE+ LP Y D L WI
Sbjct: 403 FYHGGGFIFYSAASTYFHNFCSNLANQTHSVVVSLEYRLAPEHRLPAAYEDSVEILHWIK 582
Query: 146 SHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGL---ENLPCDVKILGAI 202
+ + +PWL H +++++++ G+SAG NIA+ ++A E P ++K G I
Sbjct: 583 T-------SKDPWLTHHADYSRVYLMGESAGGNIAYTAGLRAAAIVDEIKPVNIK--GLI 735
Query: 203 IIHPYFYSANPIGSEPIIEPENN----IIHTFWHFAYPNAPFGID------NPRFNPLGE 252
+I P+F SE +E + N + + W+ + P G+D NP N
Sbjct: 736 LIQPFFGGNKRTASEIRLEKDLNLPLIVTDSMWNL---SLPLGVDRDYEYCNPTVN---G 897
Query: 253 GAPSLEKLGCSRIIVCVAGKD 273
G LEK+ V V G D
Sbjct: 898 GDKVLEKIRLFGWRVAVFGCD 960
>BG453746 weakly similar to GP|8574455|gb|A pepper esterase {Capsicum
annuum}, partial (18%)
Length = 609
Score = 117 bits (293), Expect = 6e-27
Identities = 67/182 (36%), Positives = 96/182 (51%), Gaps = 2/182 (1%)
Frame = +1
Query: 29 DGSIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPK--ITNPLSKFPILVY 86
+G+I R + P PPSL DP + SKD+ I + +RIYLP + + +K P++V+
Sbjct: 52 NGTITRLNKPPDTPPSL-DPTLPVLSKDLIINQSKGTWARIYLPHKVLDHTSNKLPLVVF 228
Query: 87 FHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISS 146
FHGG F+ S S +H A V++ SIEY LAPE+ LP Y D AL WI +
Sbjct: 229 FHGGGFILLSAASTMFHNFCLNMAKDVEVVVASIEYRLAPEHRLPAAYDDAVEALHWIKA 408
Query: 147 HSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPCDVKILGAIIIHP 206
N + WL H +++ +F+ G SAG NIA+N ++ I G I I P
Sbjct: 409 ------NLSDDWLRNHVDYSNVFLMGSSAGGNIAYNTGLRVAKAEDEISKMIKGLIXIQP 570
Query: 207 YF 208
+F
Sbjct: 571 FF 576
>TC78195 similar to GP|6092014|dbj|BAA85654.1 hsr203J homolog {Pisum
sativum}, partial (83%)
Length = 921
Score = 106 bits (265), Expect(2) = 4e-26
Identities = 72/212 (33%), Positives = 114/212 (52%), Gaps = 17/212 (8%)
Frame = +1
Query: 14 KEIVTEMGNILRIFSDGSIERPKQSPFAPPSLNDPNT-------GISSKDIQIPHNPTIS 66
K++V E+ LRI++DGS++R P + +P G++ +D+ + T +
Sbjct: 67 KKLVDEVSGWLRIYNDGSVDRTWTGPPEVTFMIEPVAPHEQFIEGVAIRDVTTTTSTTTN 246
Query: 67 S--------RIYLPKIT-NPLSKFPILVYFHGGVFMF-ESTFSKKYHEHLKTFASQANVI 116
+ R+YLP+ T K PIL++F GG F E + YH + + S + I
Sbjct: 247 TQDGSIHRARLYLPEKTPEDNKKLPILLHFQGGGFCISEPNWFMYYHVYTELVRSTRS-I 423
Query: 117 IVSIEYSLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAG 176
VS APE+ LP D +A L+W+ S + ++ +PWL EHG+F+K+F+ GDS+G
Sbjct: 424 CVSPYLRRAPEHRLPAAIDDGFATLQWLQSVAKGDVR--DPWLEEHGDFDKIFLIGDSSG 597
Query: 177 ANIAHNIAIQAGLENLPCDVKILGAIIIHPYF 208
NI H +A +AG +L V++ AI IHP F
Sbjct: 598 GNIVHEVAARAGSVDL-TPVRLARAIPIHPGF 690
Score = 28.9 bits (63), Expect(2) = 4e-26
Identities = 12/33 (36%), Positives = 19/33 (57%)
Frame = +2
Query: 245 PRFNPLGEGAPSLEKLGCSRIIVCVAGKDKLRE 277
P P+G GAP L+ L ++C+A D L++
Sbjct: 809 PSHAPMGSGAPPLDGLKLPPFLLCLAENDLLKD 907
>TC78831 similar to GP|8809707|dbj|BAA97248.1 contains similarity to unknown
protein~gb|AAF27018.1~gene_id:MQM1.21 {Arabidopsis
thaliana}, partial (50%)
Length = 1223
Score = 103 bits (257), Expect = 9e-23
Identities = 82/309 (26%), Positives = 139/309 (44%), Gaps = 22/309 (7%)
Frame = +2
Query: 36 KQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKITNPLSK---------FPILVY 86
K SP A P G+S+KD+ + + R++ P + +++ P++++
Sbjct: 191 KSSPNATPI-----NGVSTKDVTVNSENNLWFRLFTPTVAGEVTEDGGSTKTTSLPVVIF 355
Query: 87 FHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISS 146
FHGG F F S+ S Y + + + +IVS+ Y LAPE+ P+ Y D A L+++
Sbjct: 356 FHGGGFTFLSSSSNLYDAVCRRLCREISAVIVSVNYRLAPEHRYPSQYEDGEAVLRFLDE 535
Query: 147 HSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAI---QAGLENLPCDVKILGAII 203
+ L E+ + +K F+ GDSAG N+ H++A+ +AGL+N + ++G+I+
Sbjct: 536 NVT--------VLPENTDVSKCFLAGDSAGGNLVHHVAVRACKAGLQN----ICVIGSIL 679
Query: 204 IHPYFYSANPIGSEPIIEPENNII----------HTFWHFAYPNAPFGIDNPRFNPLGEG 253
I P+F G E E E ++ W P D+ N G
Sbjct: 680 IQPFF------GGEERTEAEIRLVGMPFVSVARTDWMWKVFLPEGS-DRDHGAVNVCGPN 838
Query: 254 APSLEKLGCSRIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPE 313
A L L +V V G D L + Y++ +K G K E E + H + +
Sbjct: 839 AEDLSGLDYPDTLVFVGGFDPLIDWQKRYYDWLKKCG--KKAELIEYPNMVHGFHVFPDF 1012
Query: 314 SESAKIFIQ 322
ES ++ +Q
Sbjct: 1013PESTQLIMQ 1039
>TC91859 similar to GP|8809707|dbj|BAA97248.1 contains similarity to unknown
protein~gb|AAF27018.1~gene_id:MQM1.21 {Arabidopsis
thaliana}, partial (65%)
Length = 1085
Score = 101 bits (252), Expect = 4e-22
Identities = 75/291 (25%), Positives = 135/291 (45%), Gaps = 16/291 (5%)
Frame = +2
Query: 48 PNTGISSKDIQIPHNPTISSRIYLPKIT----------NPLSKFPILVYFHGGVFMFEST 97
P G+S+KD+ + + R++ P + P++V+FHGG F + +
Sbjct: 149 PVNGVSTKDVIVNAEDNVWFRLFTPTAAVNSAGEDNTDTKTATLPVIVFFHGGGFTYLTP 328
Query: 98 FSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWISSHSNNNINNPEP 157
S Y + F + N ++VS+ Y PE+ P+ Y D A LK++ + +
Sbjct: 329 DSFAYDAVCRRFCRKINAVVVSVNYRHTPEHRYPSQYEDGEAVLKYLDEN--------KT 484
Query: 158 WLIEHGNFNKLFIGGDSAGANIAHNIAI---QAGLENLPCDVKILGAIIIHPYFYSANPI 214
L E+ + +K F+ GDSAGAN+AH++A+ +AGL +++++G + I P+F
Sbjct: 485 VLPENADVSKCFLAGDSAGANLAHHVAVRVCKAGLR----EIRVIGLVSIQPFFGGEERT 652
Query: 215 GSEPIIE--PENNIIHTFWHF-AYPNAPFGIDNPRFNPLGEGAPSLEKLGCSRIIVCVAG 271
+E +E P ++ T W + A+ D+ N G A L L +V + G
Sbjct: 653 EAEIRLEGSPLVSMARTDWMWKAFLPEGSDRDHGAVNVCGPNAEDLSGLDYPDTLVFIGG 832
Query: 272 KDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPESESAKIFIQ 322
D L + Y++ +K G K E + + H + + ES ++ +Q
Sbjct: 833 FDPLNDWQKRYYDWLKKCG--KKAELIQYPNMIHAFYIFPDLPESGQLIMQ 979
>TC76485 homologue to GP|4097684|gb|AAD00154.1| ubiquitin conjugating enzyme
{Metarhizium anisopliae}, partial (17%)
Length = 1084
Score = 99.8 bits (247), Expect = 1e-21
Identities = 45/67 (67%), Positives = 53/67 (78%)
Frame = +2
Query: 264 RIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPESESAKIFIQR 323
R+IVCVAGKDKLRERGVWY+E +K SGWKGKLE FEE+DE HVY + PESES + I+
Sbjct: 884 RMIVCVAGKDKLRERGVWYYELIKKSGWKGKLELFEEEDEDHVYHIFHPESESGQKLIKH 1063
Query: 324 LVGFVQE 330
L F+ E
Sbjct: 1064LASFLHE 1084
>TC86585 weakly similar to GP|8809631|dbj|BAA97182.1 HSR203J protein-like
protein {Arabidopsis thaliana}, partial (52%)
Length = 1226
Score = 98.6 bits (244), Expect = 3e-21
Identities = 89/315 (28%), Positives = 147/315 (46%), Gaps = 17/315 (5%)
Frame = +3
Query: 31 SIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKITNPLSKF-----PILV 85
++ R + P PSL D + + +KD+ I + R++LPK +S P++V
Sbjct: 120 TLTRNLEDPHTSPSL-DTSLSVLTKDLTINQSNQTWLRLFLPKKATNVSNLNNKLLPLIV 296
Query: 86 YFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWIS 145
+FHG F+ S S +H A ++ S++Y LAPE+ LP Y D AL I
Sbjct: 297 FFHGSGFIVLSAASTMFHNFCAEMAETVEAVVASVDYRLAPEHRLPAAYDDAMEALSLIR 476
Query: 146 SHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQA--GLENLPCDVKILGAII 203
S + + WL ++ +F+K F+ G+SAG IA++ ++ + +L +KI G I+
Sbjct: 477 S-------SDDEWLTKYVDFSKCFLMGNSAGGTIAYHAGLRVVEKMNDLE-PLKIQGLIL 632
Query: 204 IHPYFYSANPIGSEPIIEPEN----NIIHTFWHFAYPNAPFGID-NPRFNPLGEGAPSLE 258
P+F N SE +E + + W A P G++ + ++ L G E
Sbjct: 633 RQPFFGGTNRTESELRLENDPVFPLCVSDLMWELA---LPIGVNRDHEYSNLRVGNGVDE 803
Query: 259 KLGCS-----RIIVCVAGKDKLRERGVWYWEGVKNSGWKGKLEFFEEKDEGHVYQLVKPE 313
KL R++V + G D L +R E VK KG +E ++ E + + E
Sbjct: 804 KLAKIKDHEWRVLVSMNGGDPLVDRNK---ELVKLLEEKG-VEVVKDFQEDGFHGVEFFE 971
Query: 314 SESAKIFIQRLVGFV 328
AK FI+ + GF+
Sbjct: 972 LSKAKNFIEVVKGFI 1016
>TC81729 weakly similar to GP|8574455|gb|AAF77578.1| pepper esterase
{Capsicum annuum}, partial (19%)
Length = 665
Score = 98.2 bits (243), Expect = 4e-21
Identities = 61/170 (35%), Positives = 93/170 (53%), Gaps = 4/170 (2%)
Frame = +2
Query: 22 NILRIFSDGSIERPKQSPFAPPSLNDPNTGIS--SKDIQIPHNPTISSRIYLPK--ITNP 77
NI+R S+ +I R Q P+ PS +DP I+ +KDI I R++LP I+N
Sbjct: 158 NIIRT-SNNTITRLHQVPYTSPS-SDPTLPITVLTKDITINQTNKTWVRLFLPNKPISND 331
Query: 78 LSKFPILVYFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDC 137
K P++V+FHG F+ S S +H+ + V +VS+EY LAPE+ LP Y D
Sbjct: 332 HHKLPLIVFFHGSGFIVASAGSTMFHDFCVKMVDEIGVFVVSVEYRLAPEHRLPAAYDDA 511
Query: 138 WAALKWISSHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQA 187
AL WI N+ + WL + +++K ++ G SAGA IA++ ++A
Sbjct: 512 MEALFWIR-------NSDDEWLRCYVDYSKCYLMGKSAGATIAYHAGLRA 640
>TC87720 weakly similar to GP|8574455|gb|AAF77578.1| pepper esterase
{Capsicum annuum}, partial (35%)
Length = 1334
Score = 95.9 bits (237), Expect = 2e-20
Identities = 70/234 (29%), Positives = 115/234 (48%), Gaps = 13/234 (5%)
Frame = +2
Query: 31 SIERPKQSPFAPPSLNDPNTGISSKDIQIPHNPTISSRIYLPKITNPLSKF-----PILV 85
++ R + P PSL D + + +KD+ I + R++LPK +S PI+V
Sbjct: 77 TLTRYFEDPHTSPSL-DTSLPVLTKDLFINQSNQTWLRLFLPKKATNVSNLNNKLLPIIV 253
Query: 86 YFHGGVFMFESTFSKKYHEHLKTFASQANVIIVSIEYSLAPEYPLPTCYHDCWAALKWIS 145
+FHG F+ +S S +H+ A ++ S++Y LAPE+ LP Y D AL I
Sbjct: 254 FFHGSGFIVQSAASTNFHDLCVDMADTVEAVVASVDYRLAPEHRLPAAYDDAMEALSLIR 433
Query: 146 SHSNNNINNPEPWLIEHGNFNKLFIGGDSAGANIAHNIAIQAGLENLPC--DVKILGAII 203
S + + WL ++ +++K ++ G+SAGA IA++ ++ LE + +KI G I+
Sbjct: 434 S-------SQDEWLTKYVDYSKCYLMGNSAGATIAYHAGLRV-LEKVNDFEPLKIQGLIL 589
Query: 204 IHPYFYSANPIGSEPIIEPENN----IIHTFWHFAYPNAPFGID--NPRFNPLG 251
P+F N SE +E + N + W A P G+D + NP G
Sbjct: 590 RQPFFGGTNRTESELRLENDPNFPLCVSDLCWDLA---LPIGVDRNHEYCNPTG 742
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.138 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,150,275
Number of Sequences: 36976
Number of extensions: 207183
Number of successful extensions: 1254
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 1190
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1204
length of query: 330
length of database: 9,014,727
effective HSP length: 96
effective length of query: 234
effective length of database: 5,465,031
effective search space: 1278817254
effective search space used: 1278817254
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149197.10


