

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.1 + phase: 0 /pseudo
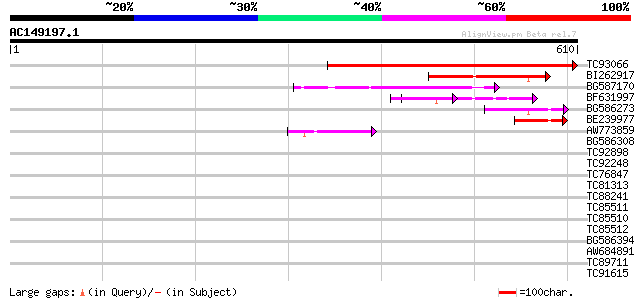
(610 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 536 e-153
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 130 1e-30
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 89 4e-18
BF631997 weakly similar to GP|18542925|gb Putative pol polyprote... 76 3e-14
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 64 2e-10
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 55 8e-08
AW773859 47 2e-05
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 39 0.006
TC92898 similar to GP|9714052|emb|CAC01296.1 sterol C-14 reducta... 37 0.022
TC92248 weakly similar to GP|22535591|dbj|BAC10766. putative DEI... 33 0.32
TC76847 homologue to SP|P08927|RUBB_PEA RuBisCO subunit binding-... 33 0.42
TC81313 similar to GP|10177964|dbj|BAB11347. urophorphyrin III m... 30 2.7
TC88241 similar to GP|11036950|gb|AAG27431.1 QM-like protein {El... 30 3.5
TC85511 homologue to SP|Q9M5M7|RL10_EUPES 60S ribosomal protein ... 30 3.5
TC85510 homologue to SP|Q9M5M7|RL10_EUPES 60S ribosomal protein ... 30 3.5
TC85512 homologue to SP|P45621|GSA_SOYBN Glutamate-1-semialdehyd... 30 3.5
BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophi... 30 3.5
AW684891 29 4.6
TC89711 weakly similar to PIR|T10725|T10725 protein kinase Xa21 ... 29 6.0
TC91615 weakly similar to GP|21593574|gb|AAM65541.1 unknown {Ara... 28 7.8
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 536 bits (1381), Expect = e-153
Identities = 262/269 (97%), Positives = 265/269 (98%), Gaps = 1/269 (0%)
Frame = +1
Query: 343 GIDKLEFCKHYVFW-KQKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIID 401
GIDKLEFCKH +F+ +KKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIID
Sbjct: 1 GIDKLEFCKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIID 180
Query: 402 DFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHG 461
DFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHG
Sbjct: 181 DFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHG 360
Query: 462 IARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPH 521
IARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPH
Sbjct: 361 IARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPH 540
Query: 522 SALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLW 581
SALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAPRA ECIFLSYASESKGYRLW
Sbjct: 541 SALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAPRAGECIFLSYASESKGYRLW 720
Query: 582 CSDPKSQKLILSRDVTFNEDALLSSGKQS 610
CSDPKSQKLILSRDVTFNEDALLSSGKQS
Sbjct: 721 CSDPKSQKLILSRDVTFNEDALLSSGKQS 807
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 130 bits (327), Expect = 1e-30
Identities = 63/134 (47%), Positives = 82/134 (61%), Gaps = 3/134 (2%)
Frame = +1
Query: 451 SDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAAS 510
+ F F + + + PQQNGVAERM RTLLER R ML AG+ + W EA
Sbjct: 25 ASF*HFVNKRVL*GNSXVAHTPQQNGVAERMNRTLLERTRAMLKTAGMA--KSFWAEAVK 198
Query: 511 TACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVND---GKLAPRAVECI 567
TAC+++NRSP + +D K P ++W G VDYS+L +FGCP Y + N KL P++ +CI
Sbjct: 199 TACYVINRSPSTVIDLKTPMEMWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCI 378
Query: 568 FLSYASESKGYRLW 581
FL YA KGY LW
Sbjct: 379 FLGYADNVKGYXLW 420
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 89.4 bits (220), Expect = 4e-18
Identities = 64/222 (28%), Positives = 98/222 (43%)
Frame = -3
Query: 306 SSSMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFST 365
SSS KD LWH LGH + ++L+ + C+ + K K F
Sbjct: 611 SSSSLNKDA--LWHARLGHPHGRALNLMLPGVVFENKN------CEACILGKHCKNVFPR 456
Query: 366 ATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKW 425
+ + D I++DLW + S +Y +T ID+ + W+ + K+ FK +
Sbjct: 455 TSTVYENCFDLIYTDLWTAPSL-SRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNF 279
Query: 426 RILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTL 485
+ V +K L +DN E+ S F +HGI + P PQQNGVA+R + L
Sbjct: 278 QAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHL 99
Query: 486 LERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFK 527
+E AR ++ A + STAC+L+N P L K
Sbjct: 98 MEVARSLMFQANV-----------STACYLINWIPTKVLRIK 6
>BF631997 weakly similar to GP|18542925|gb Putative pol polyprotein {Oryza
sativa}, partial (6%)
Length = 650
Score = 76.3 bits (186), Expect = 3e-14
Identities = 63/153 (41%), Positives = 80/153 (52%), Gaps = 6/153 (3%)
Frame = -1
Query: 422 FKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFC----TNHGIARHKTI--PRNPQQN 475
FK+W+ILVE+QTGK VK+L T+ L N FC T G +K I + +
Sbjct: 608 FKEWKILVESQTGKKVKRLQTE*RLG------NLFCRVQ*TLQGRTHYKAIYGEKYSTKE 447
Query: 476 GVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSG 535
LERARCM SNAGL R EA ST C+LVN P L V + G
Sbjct: 446 WCC*ANEYNFLERARCMFSNAGL--NRSFQAEAISTKCYLVNVLP--LLL*TVRLHLRYG 279
Query: 536 NLVDYSNLRIFGCPAYALVNDGKLAPRAVECIF 568
L++ +I GCPAY VN+GKL PR+ + +F
Sbjct: 278 -LINLLITQILGCPAYYHVNEGKLEPRSKKGLF 183
Score = 54.7 bits (130), Expect = 1e-07
Identities = 26/72 (36%), Positives = 39/72 (54%)
Frame = -2
Query: 410 YFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIP 469
+ +++K+E F + R + G+ +N LE CS++FNE C I R T+
Sbjct: 643 FMMKHKSEAFYFSRNGRFWSRVKQGRR*NVFKLNNGLEICSAEFNELCKEEHITRQYTVR 464
Query: 470 RNPQQNGVAERM 481
PQ+NGVAERM
Sbjct: 463 NTPQKNGVAERM 428
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 63.9 bits (154), Expect = 2e-10
Identities = 32/93 (34%), Positives = 56/93 (59%), Gaps = 3/93 (3%)
Frame = -2
Query: 512 ACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVND---GKLAPRAVECIF 568
AC+L+NR P L + P ++ + + +R+FGC Y LV KL R+ + +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 569 LSYASESKGYRLWCSDPKSQKLILSRDVTFNED 601
+ Y++ KGY+ C DP+++++++SRDV F E+
Sbjct: 524 IGYSTTQKGYK--CYDPEARRVLVSRDVKFIEE 432
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 55.1 bits (131), Expect = 8e-08
Identities = 29/60 (48%), Positives = 41/60 (68%), Gaps = 3/60 (5%)
Frame = -2
Query: 544 RIFGCPAYALVN-DG--KLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNE 600
RIFGC ++ ++ DG K RA++C+F+ Y+S KGYR C P S+K +SRDVTF+E
Sbjct: 456 RIFGCTSFVHIHSDGRSKFDHRALKCVFIRYSSTQKGYR--CYHPPSRKYFVSRDVTFHE 283
>AW773859
Length = 538
Score = 47.0 bits (110), Expect = 2e-05
Identities = 31/102 (30%), Positives = 46/102 (44%), Gaps = 7/102 (6%)
Frame = -3
Query: 300 TGSAAVSSSM--PKKDVT-----KLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKH 352
T SA ++SS P+ T LWH LGH++ + LLS ID+ C
Sbjct: 302 TPSATIASSQVQPQPQPTFLPQEALWHFRLGHLSNR--KLLSLHSNFPFITIDQNSVCDI 129
Query: 353 YVFWKQKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRR 394
+ + KK+ F +T+R + H D+WGP S +R
Sbjct: 128 CHYSRHKKLPFQLSTNRASKCYELFHFDIWGPFSTQSIHNQR 3
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 38.9 bits (89), Expect = 0.006
Identities = 18/49 (36%), Positives = 27/49 (54%)
Frame = -2
Query: 439 KLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLE 487
+++TDN F S+ F EFC I + PR PQ NG AE + +++
Sbjct: 670 EIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIID 524
>TC92898 similar to GP|9714052|emb|CAC01296.1 sterol C-14 reductase (FACKEL)
{Arabidopsis thaliana}, partial (55%)
Length = 689
Score = 37.0 bits (84), Expect = 0.022
Identities = 22/69 (31%), Positives = 35/69 (49%), Gaps = 10/69 (14%)
Frame = +2
Query: 25 LEWLKVLLFNLILMNLTLSFWIWKILMLKLMMKIRLFCWLSLY----------LLPTNIS 74
LE L ++LF+LIL+ + ++ M+ RLFCWL + LLP I
Sbjct: 119 LESLFLVLFSLILL----------VFIIDAMVYFRLFCWLDFFGSVPRWNLYLLLP*QIE 268
Query: 75 KKSCCMEIM 83
SCC++++
Sbjct: 269 DLSCCLQLL 295
>TC92248 weakly similar to GP|22535591|dbj|BAC10766. putative DEIH-box
RNA/DNA helicase {Oryza sativa (japonica
cultivar-group)}, partial (4%)
Length = 605
Score = 33.1 bits (74), Expect = 0.32
Identities = 12/31 (38%), Positives = 21/31 (67%)
Frame = -1
Query: 38 MNLTLSFWIWKILMLKLMMKIRLFCWLSLYL 68
+ + L W+W +L L+L +++RL WL L+L
Sbjct: 350 ITIPLWLWLWLLLWLRLRLRLRLRLWLRLWL 258
>TC76847 homologue to SP|P08927|RUBB_PEA RuBisCO subunit binding-protein
beta subunit chloroplast precursor (60 kDa chaperonin
beta subunit), partial (96%)
Length = 2096
Score = 32.7 bits (73), Expect = 0.42
Identities = 25/105 (23%), Positives = 49/105 (45%), Gaps = 23/105 (21%)
Frame = +1
Query: 30 VLLFNLILMNLTLSFWIWKILMLKLMMK------IRLFCWLSLYLLP------------- 70
VLL L++++L L W W+I + L++ R WL++ LLP
Sbjct: 409 VLLRLLMMVSLLLERWSWRIRLRILVLNW*DRRLPRQMTWLAMELLPLLFWLKVLLQKVL 588
Query: 71 --TNISKKSCCMEIMI--PYPLKMLNLICCPKKVFILKFILWTRV 111
+ + C +++ L +LNL C K++ I+ +++W ++
Sbjct: 589 RLLQLVQTLCLSHVVLRRQRKLLLLNLN*CQKRLRIVNWLMWQQL 723
>TC81313 similar to GP|10177964|dbj|BAB11347. urophorphyrin III methylase
{Arabidopsis thaliana}, partial (71%)
Length = 1121
Score = 30.0 bits (66), Expect = 2.7
Identities = 13/32 (40%), Positives = 20/32 (61%)
Frame = +1
Query: 25 LEWLKVLLFNLILMNLTLSFWIWKILMLKLMM 56
LE L+VL+ ++ L + FWIW ++ML M
Sbjct: 517 LELLRVLICCCMIGWLVMMFWIWLVIMLNFCM 612
>TC88241 similar to GP|11036950|gb|AAG27431.1 QM-like protein {Elaeis
guineensis}, partial (95%)
Length = 954
Score = 29.6 bits (65), Expect = 3.5
Identities = 10/22 (45%), Positives = 16/22 (72%)
Frame = +1
Query: 341 KQGIDKLEFCKHYVFWKQKKVS 362
K+G+D+ FC H V W+++ VS
Sbjct: 199 KKGVDEFPFCVHLVSWEKENVS 264
>TC85511 homologue to SP|Q9M5M7|RL10_EUPES 60S ribosomal protein L10. [Leafy
spurge] {Euphorbia esula}, partial (97%)
Length = 891
Score = 29.6 bits (65), Expect = 3.5
Identities = 10/22 (45%), Positives = 16/22 (72%)
Frame = +2
Query: 341 KQGIDKLEFCKHYVFWKQKKVS 362
K+G+D+ FC H V W+++ VS
Sbjct: 173 KKGVDEFPFCVHLVSWEKENVS 238
>TC85510 homologue to SP|Q9M5M7|RL10_EUPES 60S ribosomal protein L10. [Leafy
spurge] {Euphorbia esula}, partial (97%)
Length = 904
Score = 29.6 bits (65), Expect = 3.5
Identities = 10/22 (45%), Positives = 16/22 (72%)
Frame = +2
Query: 341 KQGIDKLEFCKHYVFWKQKKVS 362
K+G+D+ FC H V W+++ VS
Sbjct: 161 KKGVDEFPFCVHLVSWEKENVS 226
>TC85512 homologue to SP|P45621|GSA_SOYBN Glutamate-1-semialdehyde 2
1-aminomutase chloroplast precursor (EC 5.4.3.8) (GSA),
partial (91%)
Length = 2272
Score = 29.6 bits (65), Expect = 3.5
Identities = 10/22 (45%), Positives = 16/22 (72%)
Frame = -2
Query: 341 KQGIDKLEFCKHYVFWKQKKVS 362
K+G+D+ FC H V W+++ VS
Sbjct: 483 KKGVDEFPFCVHLVSWEKENVS 418
>BG586394 similar to GP|7295463|gb| CG4835 gene product {Drosophila
melanogaster}, partial (2%)
Length = 734
Score = 29.6 bits (65), Expect = 3.5
Identities = 15/46 (32%), Positives = 30/46 (64%), Gaps = 3/46 (6%)
Frame = -2
Query: 28 LKVLLFNLILM---NLTLSFWIWKILMLKLMMKIRLFCWLSLYLLP 70
+++LLF LM +L F +++++ L+L++ L W+SL+L+P
Sbjct: 535 VQLLLFYFFLML**SLEYLFLVFQLVSLQLLLFPFLISWVSLFLIP 398
>AW684891
Length = 539
Score = 29.3 bits (64), Expect = 4.6
Identities = 17/84 (20%), Positives = 43/84 (50%)
Frame = -3
Query: 396 MMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNE 455
++T+I + K ++ F+ F K + + Q K++++L ++ + + ++
Sbjct: 372 VLTLIQIYGXKWFLAFIHNSTSIT*LFLK-KHEIWFQFVKSIERLCSNKGKVYVDQNLSQ 196
Query: 456 FCTNHGIARHKTIPRNPQQNGVAE 479
+ +G+++ +PQQNGV+E
Sbjct: 195 YLEENGVSQELKCVNSPQQNGVSE 124
>TC89711 weakly similar to PIR|T10725|T10725 protein kinase Xa21 (EC
2.7.1.-) A1 receptor type - long-staminate rice,
partial (7%)
Length = 1391
Score = 28.9 bits (63), Expect = 6.0
Identities = 9/22 (40%), Positives = 16/22 (71%)
Frame = -2
Query: 363 FSTATHRTKGILDYIHSDLWGP 384
F ++T + +L+ IH+D+WGP
Sbjct: 901 FKSSTSHAQEVLELIHTDVWGP 836
>TC91615 weakly similar to GP|21593574|gb|AAM65541.1 unknown {Arabidopsis
thaliana}, partial (50%)
Length = 834
Score = 28.5 bits (62), Expect = 7.8
Identities = 15/41 (36%), Positives = 23/41 (55%)
Frame = +3
Query: 31 LLFNLILMNLTLSFWIWKILMLKLMMKIRLFCWLSLYLLPT 71
LLFN + T F W I++L L + +RLF L ++ + T
Sbjct: 549 LLFNPPPLRFTAFFPRWMIIVLLLDLNVRLFMRLLIFFVAT 671
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,376,601
Number of Sequences: 36976
Number of extensions: 334697
Number of successful extensions: 2376
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 2337
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2364
length of query: 610
length of database: 9,014,727
effective HSP length: 102
effective length of query: 508
effective length of database: 5,243,175
effective search space: 2663532900
effective search space used: 2663532900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149197.1


