

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.9 + phase: 0 /pseudo
(1214 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
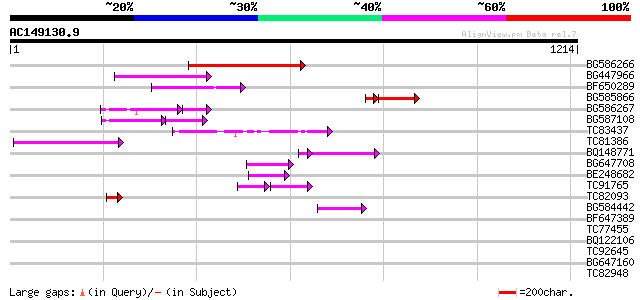
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 204 2e-52
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 148 1e-35
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 137 2e-32
BG585866 130 5e-32
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 99 2e-28
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 77 1e-26
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 105 1e-22
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 93 7e-19
BQ148771 81 1e-16
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 85 1e-16
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 66 7e-11
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 57 4e-08
TC82093 44 5e-04
BG584442 43 6e-04
BF647389 42 0.001
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 39 0.012
BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2... 38 0.027
TC92645 37 0.046
BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrio... 36 0.079
TC82948 36 0.10
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 204 bits (519), Expect = 2e-52
Identities = 107/251 (42%), Positives = 153/251 (60%), Gaps = 1/251 (0%)
Frame = -3
Query: 383 DFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPGRGTADNSIILQEILHFMKR 442
++R I+ CNT YKII K+L R++P L ++I P QS+F+PGR +DN +I +ILH++++
Sbjct: 772 EYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYLRQ 593
Query: 443 SKRKKGY-VAFKLDLEKAFDNVNWDFLNSCLLDFGFPDIIVKLIMHCVSSANYSLLWNGN 501
S KK +A K D+ KA+D + W+FL L GF I + IM CVS+ +YS L NG
Sbjct: 592 SGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLINGG 413
Query: 502 KMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHLLFA 561
P+ GLRQGDPLSPYLFILC E LS Q A+ +G+ + + + P I+HLLFA
Sbjct: 412 PQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLLFA 233
Query: 562 DDVLLFTKAKSSQLQFITNLFDRFSRASGLKINISKSRAYYSSGTPNGKINNLTAISGIQ 621
DD + F K+ +S + ++ D++ ASG IN +KS +SS T I+ + I
Sbjct: 232 DDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELKIA 53
Query: 622 STTTLGKYLGF 632
GKYLG+
Sbjct: 52 KEGGTGKYLGY 20
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 148 bits (373), Expect = 1e-35
Identities = 83/209 (39%), Positives = 114/209 (53%)
Frame = +2
Query: 224 WVKLGDKNTAFFHAQTVIRRKWNKIHKLQLPNGISTSDSNILQEEALKYFKKFFCGSQIP 283
W+K GDKNT FFH++ RRK N+I KL+ G ++ + YF F S
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPT 184
Query: 284 YSRFFNEGRHPALDDTGKTSLTSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIV 343
E L T++EV A+N M P KAPGPDG +FF++YWHIV
Sbjct: 185 AIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIV 364
Query: 344 GDDVFHLVRSAFLTGHFDPAISNTLIALIPKIDSPNTYKDFRPISLCNTLYKIITKVLVH 403
G +V +V ++ T I LIPK +PNT KD+RPISLCN + KIITKV+ +
Sbjct: 365 GKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIAN 544
Query: 404 RLRPFLNNLIGPYQSSFLPGRGTADNSII 432
R++ L ++I QS+F+ GR DN++I
Sbjct: 545 RVKQTLPDVIDVEQSAFVQGRLITDNALI 631
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 137 bits (346), Expect = 2e-32
Identities = 77/205 (37%), Positives = 118/205 (57%), Gaps = 3/205 (1%)
Frame = +3
Query: 304 LTSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTGHFDPA 363
L S T EV AL SM KAPG DG++ FFK W+I+GD V + F TG
Sbjct: 12 LCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKI 191
Query: 364 ISNTLIALIPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPG 423
I+ T + L+PK + + K+FRPI+ C+ +YKII+K+L R++ LN+++ QS+F+ G
Sbjct: 192 INCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKG 371
Query: 424 RGTADNSIILQEILHFMKRSKRKKGY---VAFKLDLEKAFDNVNWDFLNSCLLDFGFPDI 480
R DN I+ E++ +S +KG K+DL KA+D+ W F+ +L+ GFP
Sbjct: 372 RVIFDNIILSHELV----KSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYK 539
Query: 481 IVKLIMHCVSSANYSLLWNGNKMPP 505
V +M +++A+Y+ NG+ P
Sbjct: 540 FVNWVMAXLTTASYTFNXNGDLTXP 614
>BG585866
Length = 828
Score = 130 bits (327), Expect(2) = 5e-32
Identities = 58/89 (65%), Positives = 72/89 (80%)
Frame = +3
Query: 789 IIRAKDILKTGYSWRAGAGTSSFWFSNWSSHGYLGSLVPIIDIHDIHLTVKDVLTSVGQH 848
IIR K++LK+GY+WRAG+G SSFW++NWSS G LG+ P +DIHD+HLTVKDV T+ GQH
Sbjct: 84 IIRDKNVLKSGYTWRAGSGNSSFWYTNWSSLGLLGTQAPFVDIHDLHLTVKDVFTTGGQH 263
Query: 849 TNVLYTNLPQGIAETINNSHMRFNANIDD 877
T LYT LP IAE INN+H+ FNA+I D
Sbjct: 264 TQSLYTILPTDIAEVINNTHLNFNASIGD 350
Score = 26.9 bits (58), Expect(2) = 5e-32
Identities = 15/28 (53%), Positives = 17/28 (60%), Gaps = 1/28 (3%)
Frame = +1
Query: 763 LAKKYSSGTSLLE-ANVNSNSSPSWFSI 789
L YSSG + L A + NSSPSW SI
Sbjct: 1 LFNNYSSGHNFLHIAAAHRNSSPSWSSI 84
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 99.0 bits (245), Expect(2) = 2e-28
Identities = 59/180 (32%), Positives = 90/180 (49%), Gaps = 5/180 (2%)
Frame = +3
Query: 195 TLLEKELQDEYNHILFQEEMLWYQKSREQWVKLGDKNTAFFHAQTVIRRKWNKIHKLQLP 254
+LL EL +EY++ EE+ W QKSR W++ GD+NT FFHA T RR N+I L
Sbjct: 84 SLLRSELNEEYHN----EEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSL--- 242
Query: 255 NGISTSDSNILQEEAL-----KYFKKFFCGSQIPYSRFFNEGRHPALDDTGKTSLTSPIT 309
I D EE L +FK + + + + + L + I+
Sbjct: 243 --IDDDDKEWFVEEDLGRLADSHFKLLYSSEDVGITLEDWNSIPAIVTEEQNAQLMAQIS 416
Query: 310 KKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAFLTGHFDPAISNTLI 369
++EV A+ + P+K PGPDG + FF+Q+W +GDD+ + + TG + I+ T I
Sbjct: 417 REEVREAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEGINKTNI 596
Score = 46.6 bits (109), Expect(2) = 2e-28
Identities = 26/62 (41%), Positives = 37/62 (58%)
Frame = +1
Query: 371 LIPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRPFLNNLIGPYQSSFLPGRGTADNS 430
L+PK +FRPISLCN YKI++KVL RL+ L +I Q++F + +DN
Sbjct: 601 LVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNI 780
Query: 431 II 432
+I
Sbjct: 781 LI 786
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 77.4 bits (189), Expect(2) = 1e-26
Identities = 40/89 (44%), Positives = 53/89 (58%)
Frame = +3
Query: 335 FFKQYWHIVGDDVFHLVRSAFLTGHFDPAISNTLIALIPKIDSPNTYKDFRPISLCNTLY 394
FF+ WHI+ D+ +V S +G D ++ T I LIPK P + RPISLCN Y
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 395 KIITKVLVHRLRPFLNNLIGPYQSSFLPG 423
KII+KVL RL+ L +LI QS+F+ G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
Score = 62.0 bits (149), Expect(2) = 1e-26
Identities = 45/140 (32%), Positives = 66/140 (47%)
Frame = +2
Query: 197 LEKELQDEYNHILFQEEMLWYQKSREQWVKLGDKNTAFFHAQTVIRRKWNKIHKLQLPNG 256
L+++LQ+ Y EE W QKSR W GD N F+HA T R N+I L +G
Sbjct: 8 LKEKLQEAYK----DEEDYWQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDG 175
Query: 257 ISTSDSNILQEEALKYFKKFFCGSQIPYSRFFNEGRHPALDDTGKTSLTSPITKKEVFAA 316
++ +++ A+ YF+ F + F + ++ L T++EV A
Sbjct: 176 NWITEEQGVEKVAVDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLATEEEVRLA 355
Query: 317 LNSMKPYKAPGPDGFHCIFF 336
L M P KAPGPDG + F
Sbjct: 356 LFIMHPEKAPGPDGMTTLLF 415
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 105 bits (262), Expect = 1e-22
Identities = 105/360 (29%), Positives = 159/360 (44%), Gaps = 16/360 (4%)
Frame = +2
Query: 348 FHLVRSAFLTGHFDPAISNTLIALIPKIDSPNTYKDFRPISLCNTLYKIITKVLVHRLRP 407
FH R F I++T IALIPK+D+P DFRPISL +LYKI+ K+L +RLR
Sbjct: 20 FHRNRKLF------KGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRV 181
Query: 408 FLNNLIGPYQSSFLPGRGTADNSIILQEILHFMKRSKRKKGYVAFKLDLEKAFDNVNWDF 467
+ ++I QS+F+ R + + Q L R+ R K F + W
Sbjct: 182 VIGSVISDAQSAFVKNRQILEMVFL*QMRLWMRLRN*R------------KIFCCLRWIL 325
Query: 468 LNSCLLDFGFPDIIV-----------KLIMHCVSSANYSLLWNGNKMPPFKPTHGLRQGD 516
L G I+ K I CVS+A S+L NG+ PT+ L +
Sbjct: 326 KRLITLSIGLIWILF*VGMSFLVLWRKWIKECVSTATTSVLVNGS------PTNVLMK-S 484
Query: 517 PLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHLLFADDVLLFTKAKSSQLQ 576
+ LF + S + + V+ +SHL FA+D LL + ++
Sbjct: 485 LVQTQLF----TRYSFGVVNPVV---------------VSHLQFANDTLLLETKNWANIR 607
Query: 577 FITNLFDRFSRASGLKINISKSRAYYSSGTPNGKINNLTAISGIQSTTTLGK----YLGF 632
+ F SGLK+N KS + P+ + +S +GK YLG
Sbjct: 608 ALRAALVIF*AMSGLKVNFHKSGLVCVNIAPSWLSEAASVLS-----WKVGKVPFLYLGM 772
Query: 633 PMLQGRPKRSDF-NFILEKMQTRLASWKNRLLNRTGRLTLATSVLSTIPTYYMQINWLPQ 691
P ++G +R F I+ +++ RL W +R L+ GRL L SVL+++ Y + + L Q
Sbjct: 773 P-IEGNSRRLSFWEPIVNRIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYALPSSKLHQ 949
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 92.8 bits (229), Expect = 7e-19
Identities = 71/244 (29%), Positives = 109/244 (44%), Gaps = 9/244 (3%)
Frame = -1
Query: 9 WMLIGDFNETHLPSEQRGGTFHHNRAAT--FSNFMNNCNLLDLTTTGGRFTWHKNNNGIR 66
W IGDFN T L S + G+ R F + + NL L T G FTW G
Sbjct: 741 WCCIGDFN-TILGSHEHQGSHTPARLPMLDFQQWSDVNNLFHLPTRGSAFTWTNGRRGRN 565
Query: 67 ILSKKLDRGMANVDWRLSFPEAFVEVLCRLHSDHNPLLLRFGGLPLTRGPRPFRFEAAWI 126
K+LDR + N + L +L SDH P+L + + F+F W
Sbjct: 564 NTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILFELQTQNI-QFSSSFKFMKMWS 388
Query: 127 DHYDYGNVVKRSWSTHTHN-PTASLIKVMENSI----IFNHDVFGNIFQRKSRVEWRLKG 181
H D N++K+ W+ P L + ++N ++N + FGN+ + L
Sbjct: 387 AHPDCINIIKQCWANQVVGCPMFVLNQKLKNLKEVLKVWNKNTFGNVHSQVDNAYKELDD 208
Query: 182 VQSYLERVDSYRHTLLEKELQDEYN--HILFQEEMLWYQKSREQWVKLGDKNTAFFHAQT 239
+Q ++ + Y L+++E + N L EE+ W++KS+ W GD+NTAFFH
Sbjct: 207 IQVKIDSI-GYSDVLMDQEKAAQLNLESALNIEEVFWHEKSKVNWHCEGDRNTAFFHRVA 31
Query: 240 VIRR 243
I+R
Sbjct: 30 KIKR 19
>BQ148771
Length = 680
Score = 81.3 bits (199), Expect(2) = 1e-16
Identities = 46/154 (29%), Positives = 81/154 (51%), Gaps = 2/154 (1%)
Frame = -3
Query: 640 KRSDFNFILEKMQTRLASWKNRLLNRTGRLTLATSVLSTIPTYYMQINWLPQNICDSIDQ 699
K+S F+ + ++ LA+WK L+ R+TLA SV+ +P Y M +P+ + I +
Sbjct: 615 KKSRFS-VYYQVHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQK 439
Query: 700 TARNFLWKGSN-NKGIHLVNWKTITRPKSIGGLGIRSARDANICLLGKLVWDMVQSTNKL 758
R F+W + ++ H V W+T+++PK+I GLG+R N + KL W + +N L
Sbjct: 438 LQRKFVWGDTEVSRRYHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSNSL 259
Query: 759 WVNLLAKKYSSGTSLLEANVNSNSSPS-WFSIIR 791
++ KY SL E + + S W ++++
Sbjct: 258 CTEVMRGKYQRSESLEEIFLEKPTDSSLWKALVK 157
Score = 24.3 bits (51), Expect(2) = 1e-16
Identities = 10/30 (33%), Positives = 16/30 (53%)
Frame = -2
Query: 618 SGIQSTTTLGKYLGFPMLQGRPKRSDFNFI 647
+G + T+LGKY G P + K F+ +
Sbjct: 679 AGFRXATSLGKYXGVPC*EEHQKEPIFSIL 590
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 85.1 bits (209), Expect = 1e-16
Identities = 44/101 (43%), Positives = 57/101 (55%)
Frame = +1
Query: 508 PTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHLLFADDVLLF 567
P GLRQGDPLSPYLFILC LS ++ + + I + P+I+HLLFADD LLF
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 568 TKAKSSQLQFITNLFDRFSRASGLKINISKSRAYYSSGTPN 608
+A ++ I + + ASG +N KS YS PN
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPN 306
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 66.2 bits (160), Expect = 7e-11
Identities = 34/88 (38%), Positives = 51/88 (57%)
Frame = +3
Query: 511 GLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSWEPIHIINDGPQISHLLFADDVLLFTKA 570
GL+QGDPL+P+LF+L E +S +++AV + ++ + G ++SHL +ADD L
Sbjct: 27 GLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIGMP 206
Query: 571 KSSQLQFITNLFDRFSRASGLKINISKS 598
L + L F ASGLK+N KS
Sbjct: 207 TVDNLWTLKALLQGFEMASGLKVNFHKS 290
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 57.0 bits (136), Expect = 4e-08
Identities = 29/69 (42%), Positives = 40/69 (57%)
Frame = +2
Query: 488 CVSSANYSLLWNGNKMPPFKPTHGLRQGDPLSPYLFILCMEKLSVAIQDAVLQGSWEPIH 547
CV S +Y +L N + + P P+ GL+QGD LSPY+FI+C+E LS I A +G
Sbjct: 104 CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTS 283
Query: 548 IINDGPQIS 556
I P +S
Sbjct: 284 I*RGAPPVS 310
Score = 43.1 bits (100), Expect = 6e-04
Identities = 27/90 (30%), Positives = 42/90 (46%)
Frame = +3
Query: 558 LLFADDVLLFTKAKSSQLQFITNLFDRFSRASGLKINISKSRAYYSSGTPNGKINNLTAI 617
LLF L + + Q + N+ + SG I++ KS Y S P+ ++T I
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 618 SGIQSTTTLGKYLGFPMLQGRPKRSDFNFI 647
G+Q KYLG P + GR + + F+ I
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTFSSI 548
>TC82093
Length = 569
Score = 43.5 bits (101), Expect = 5e-04
Identities = 19/33 (57%), Positives = 23/33 (69%)
Frame = -1
Query: 208 ILFQEEMLWYQKSREQWVKLGDKNTAFFHAQTV 240
+L Q EMLWY KS+ WVK G +NT FFH + V
Sbjct: 476 LLSQNEMLWY*KSQ*NWVKYGSRNTWFFHNEDV 378
>BG584442
Length = 775
Score = 43.1 bits (100), Expect = 6e-04
Identities = 28/108 (25%), Positives = 52/108 (47%), Gaps = 2/108 (1%)
Frame = +1
Query: 659 KNRLLNRTGRLTLATSVLSTIPTYYMQINWLPQNICDSIDQTARNFLWK--GSNNKGIHL 716
+N+ L++ + L +I +Y M I L + D I++ F W G N KG+H
Sbjct: 397 RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHVGENRKGMHW 576
Query: 717 VNWKTITRPKSIGGLGIRSARDANICLLGKLVWDMVQSTNKLWVNLLA 764
++ + + K+ GG+G NI +LGK V + + L++ ++
Sbjct: 577 MS*EKLFVHKNYGGMGFTDFTTFNIPMLGKQV*SFLLNRTTLFLEKIS 720
>BF647389
Length = 404
Score = 42.0 bits (97), Expect = 0.001
Identities = 34/124 (27%), Positives = 57/124 (45%), Gaps = 1/124 (0%)
Frame = +2
Query: 628 KYLGFPMLQGRPKRSDFNFILEKMQTRLASWKNRLLNRTGRLTLATSVLSTIPTYYMQIN 687
+YLG P+ K+ ++K+ R+ + ++LL+ G L L VL + Y+ Q+
Sbjct: 41 RYLGVPLSS---KKLYVIQRVKKIICRIEN*SSKLLSYAGSLQLIKIVLFGVQPYWSQVF 211
Query: 688 WLPQNICDSIDQTARNFLWKGSNNKGIH-LVNWKTITRPKSIGGLGIRSARDANICLLGK 746
LP + I T R FL G + L+ + I PK+ GG + + N + K
Sbjct: 212 VLP*KVIKLIQTTCRIFL*TGKSGTSKRALIAREHICLPKTAGGWNVIDLKVXNQTAICK 391
Query: 747 LVWD 750
L W+
Sbjct: 392 LXWN 403
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 38.9 bits (89), Expect = 0.012
Identities = 18/48 (37%), Positives = 25/48 (51%)
Frame = -3
Query: 725 PKSIGGLGIRSARDANICLLGKLVWDMVQSTNKLWVNLLAKKYSSGTS 772
P+ GGLG+R R N+ LL K W ++Q + LW +L Y S
Sbjct: 963 PRCKGGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIYGPRVS 820
>BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2 finger
protein rha1a. {Trypanosoma brucei}, partial (18%)
Length = 693
Score = 37.7 bits (86), Expect = 0.027
Identities = 47/151 (31%), Positives = 60/151 (39%)
Frame = -3
Query: 1033 SRVTSFSRIQLAW*NGIKETINVTFSM*TAVVSELQFELDSEVFFETTLVVTYLVTPVLS 1092
S +T F + W*NGI T V FSM T VV DS T LV+P
Sbjct: 613 SCITPFLKRPTPW*NGIATTTLVIFSMLTGVVLATLSLPDSTASLGTLRDYLTLVSPGTL 434
Query: 1093 RNRQTSC*LNSLLFIKA*SWQLKWE*KS*LATRTRFLPLISSQEQLQNTILTLS*YKT*R 1152
R T NS+ K WE AT T L+SS + + + +KT*R
Sbjct: 433 PTRLTFFWPNSMPSFKVCE*FQIWEYLILCATSTLSTMLVSSMDLP*SFMSMPLSFKT*R 254
Query: 1153 TSYPHIISQSIIVSGKGINARTTWRNWERHR 1183
TS + + I A+ + WER R
Sbjct: 253 TSSLPARLLFFTLFVREITAQIFLKCWERLR 161
>TC92645
Length = 561
Score = 37.0 bits (84), Expect = 0.046
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 2/90 (2%)
Frame = +3
Query: 296 LDDTGKTSLTSPITKKEVFAALNSMKPYKAPGPDGFHCIFFKQYWHIVGDDVFHLVRSAF 355
+ D LT+P + E++ A+ M KA G DG + + ++ ++VG D + V
Sbjct: 282 IGDQNNCMLTAPFSVDEIWTAIFQMHHDKAFGLDGLNLAVYYRF*NLVGGDT-NYVGLYS 458
Query: 356 LTGHFDPA--ISNTLIALIPKIDSPNTYKD 383
L G A T I L+PK DSP+T D
Sbjct: 459 LVG*R*VA*LYWGTNIVLVPKCDSPSTMWD 548
>BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrioides}, partial
(2%)
Length = 780
Score = 36.2 bits (82), Expect = 0.079
Identities = 44/129 (34%), Positives = 53/129 (40%), Gaps = 1/129 (0%)
Frame = +2
Query: 1057 FSM*TAVVSELQFELDSEVFFETTLVVTYLVTPVLSRNRQTSC*LNSLLFIKA*SWQLKW 1116
F M S F D V F T +T V V + SC LN LFIK *+WQ
Sbjct: 329 F*MSKEAASVFLFTHDLVVLFATI*ALT*QVFRVSYPFIKISCLLN*QLFIKV*NWQSP* 508
Query: 1117 E*KS*LATRTRFLPLISSQE-QLQNTILTLS*YKT*RTSYPHIISQSIIVSGKGINARTT 1175
K T + ISS+ L T + S + T TS IS II + IN R +
Sbjct: 509 ILKKWFVILTLYSLSISSRRISLSTTFMRFS-FITSNTS*ALGISLFIIHLERAINVRIS 685
Query: 1176 WRNWERHRM 1184
W N E M
Sbjct: 686 WSNLEPRLM 712
>TC82948
Length = 705
Score = 35.8 bits (81), Expect = 0.10
Identities = 18/38 (47%), Positives = 22/38 (57%)
Frame = +3
Query: 717 VNWKTITRPKSIGGLGIRSARDANICLLGKLVWDMVQS 754
V+W+ + RP G LGIRS N L KL WDM+ S
Sbjct: 267 VSWEKVCRPIKEGSLGIRSLSKLNEALNLKLCWDMMIS 380
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.143 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,318,800
Number of Sequences: 36976
Number of extensions: 659351
Number of successful extensions: 3845
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 2827
Number of HSP's successfully gapped in prelim test: 136
Number of HSP's that attempted gapping in prelim test: 958
Number of HSP's gapped (non-prelim): 3052
length of query: 1214
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1107
effective length of database: 5,058,295
effective search space: 5599532565
effective search space used: 5599532565
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149130.9


