

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.7 - phase: 0 /pseudo
(359 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
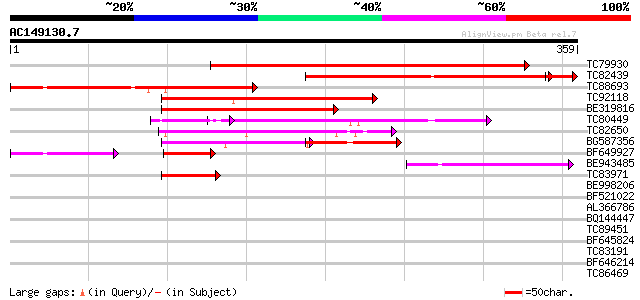
Score E
Sequences producing significant alignments: (bits) Value
TC79930 weakly similar to PIR|T04241|T04241 hypothetical protein... 286 9e-78
TC82439 similar to GP|17065050|gb|AAL32679.1 putative protein {A... 215 5e-59
TC88693 similar to GP|17065050|gb|AAL32679.1 putative protein {A... 174 4e-44
TC92118 similar to PIR|T48532|T48532 hypothetical protein T22P22... 150 1e-36
BE319816 similar to PIR|G96707|G96 hypothetical protein T2E12.6 ... 112 3e-25
TC80449 weakly similar to GP|15293267|gb|AAK93744.1 unknown prot... 110 1e-24
TC82650 similar to GP|13374866|emb|CAC34500. putative protein {A... 82 2e-16
BG587356 weakly similar to GP|22535532|db hypothetical protein~s... 81 5e-16
BF649927 similar to GP|17065506|gb| Unknown protein {Arabidopsis... 66 2e-11
BE943485 similar to PIR|T48532|T485 hypothetical protein T22P22.... 54 7e-08
TC83971 weakly similar to GP|23315108|gb|AAN20289.1 Sequence 273... 50 1e-06
BE998206 weakly similar to GP|20804766|dbj contains ESTs AU09611... 39 0.002
BF521022 homologue to GP|17944908|gb LP04958p {Drosophila melano... 30 1.9
AL366786 similar to PIR|C86405|C86 probable sphingosine-1-phosph... 30 1.9
BQ144447 GP|12697999|d KIAA1727 protein {Homo sapiens}, partial ... 28 5.5
TC89451 weakly similar to GP|21536959|gb|AAM61300.1 unknown {Ara... 28 7.1
BF645824 similar to PIR|T07142|T07 beta-glucan-elicitor receptor... 28 7.1
TC83191 similar to GP|6714272|gb|AAF25968.1| F6N18.8 {Arabidopsi... 27 9.3
BF646214 similar to GP|8843779|dbj| gene_id:MZN1.3~unknown prote... 27 9.3
TC86469 similar to GP|21464654|emb|CAD30328. acid phosphatase {L... 27 9.3
>TC79930 weakly similar to PIR|T04241|T04241 hypothetical protein F14M19.150
- Arabidopsis thaliana, partial (53%)
Length = 1154
Score = 286 bits (732), Expect = 9e-78
Identities = 130/202 (64%), Positives = 163/202 (80%)
Frame = +2
Query: 128 VYVHASKAKPVHVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDS 187
VYVHASK KP+HVS YFV RDI S+ + WG++S+VEAERRLLANAL DP+NQHFVLLSDS
Sbjct: 2 VYVHASKEKPIHVSPYFVGRDIHSEPVTWGRISMVEAERRLLANALLDPDNQHFVLLSDS 181
Query: 188 CVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKR 247
C+P+ F+++++YL+ T SF+DS+ D GP GNGRY E MLPEVE KDFR G+QWFS+KR
Sbjct: 182 CIPVRRFDFVYNYLLLTSVSFIDSYVDTGPHGNGRYVERMLPEVEKKDFRKGSQWFSMKR 361
Query: 248 QHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRS 307
QHA+ +MAD LY++KF+ C + +NC DEHYLPTFF ++DP GIA WSVTYVD S
Sbjct: 362 QHALIIMADSLYFNKFKHHCRPNMEGNRNCYSDEHYLPTFFNMLDPGGIANWSVTYVDWS 541
Query: 308 EQKRHPKSYRTQDITYELLKNI 329
E K HP+S+R +DITY+L+K I
Sbjct: 542 EGKWHPRSFRARDITYKLMKKI 607
>TC82439 similar to GP|17065050|gb|AAL32679.1 putative protein {Arabidopsis
thaliana}, partial (47%)
Length = 1019
Score = 215 bits (548), Expect(2) = 5e-59
Identities = 103/157 (65%), Positives = 123/157 (77%)
Frame = +1
Query: 188 CVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKR 247
C ++YI++YLM+T+ S+VD F+DPGP GNGRYS MLPEVE+KDFR GAQWFS+KR
Sbjct: 22 CTIYIIYDYIYNYLMHTNISYVDCFKDPGPHGNGRYSHRMLPEVEVKDFRKGAQWFSMKR 201
Query: 248 QHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRS 307
QHAV VMAD+LYYSKF+A C Q ++GKNCI DEHYLPTFF IVDP GIA WSVT+VD S
Sbjct: 202 QHAVIVMADYLYYSKFRAYC-QPGLEGKNCIADEHYLPTFFQIVDPGGIANWSVTHVDWS 378
Query: 308 EQKRHPKSYRTQDITYELLKNIKLHNFHLFLTERSTK 344
E+K HPKSYR D+TYELLKNI + + +T K
Sbjct: 379 ERKWHPKSYRDHDVTYELLKNITSVDVSVHVTSDEKK 489
Score = 30.4 bits (67), Expect(2) = 5e-59
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +3
Query: 340 ERSTKMDLFLEWISEAVLPI 359
ERS K+ L +EW SEA+LPI
Sbjct: 486 ERSAKLALLMEWDSEAMLPI 545
>TC88693 similar to GP|17065050|gb|AAL32679.1 putative protein {Arabidopsis
thaliana}, partial (26%)
Length = 952
Score = 174 bits (442), Expect = 4e-44
Identities = 100/171 (58%), Positives = 119/171 (69%), Gaps = 14/171 (8%)
Frame = +1
Query: 1 MKTAKFWRL-SMGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTS- 58
MK+ K WRL SMGDM+IL GS HRP MKKPMWI+VLV F+ +FL AY+Y Q+++S
Sbjct: 445 MKSDKDWRLGSMGDMQILPGSR--HRPPMKKPMWIIVLVLFVCVFLIIAYMYPPQSSSSA 618
Query: 59 CNMFYSKPCIIDISVLAVSHIPVSVYIE-----RGITLDMPQNP-------KIAFMFLTP 106
C +F S+ C L + P Y + R + D+ +P KIAFMFL+P
Sbjct: 619 CYIFSSRGCKRFSDWLPPA--PAREYTDEEIASRVVIKDILTSPPVVSKKSKIAFMFLSP 792
Query: 107 GSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQLVWG 157
GSLP EKLWDNFFQGHEGKFSVYVHASK+KP HVSRYFVNRDIRS Q+VWG
Sbjct: 793 GSLPLEKLWDNFFQGHEGKFSVYVHASKSKPDHVSRYFVNRDIRSGQVVWG 945
>TC92118 similar to PIR|T48532|T48532 hypothetical protein T22P22.120 -
Arabidopsis thaliana, partial (51%)
Length = 1044
Score = 150 bits (378), Expect = 1e-36
Identities = 71/139 (51%), Positives = 96/139 (68%), Gaps = 2/139 (1%)
Frame = +1
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHV--SRYFVNRDIRSDQL 154
PKIAFMFLT G LP +W+ F +GHE ++SVYVH + + H S F R I S
Sbjct: 622 PKIAFMFLTKGPLPLAPVWERFLKGHESRYSVYVHPTPSYQAHFPPSSVFYKRQIPSQIA 801
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG+MS+ +AERRLLANAL D +N+ F+LLS+SC+PLY F+ +++YLM + SFV +F D
Sbjct: 802 EWGRMSMCDAERRLLANALLDISNERFILLSESCIPLYKFSLVYNYLMKSKFSFVGAFDD 981
Query: 215 PGPVGNGRYSEHMLPEVEI 233
PGP G RY+ +M P V++
Sbjct: 982 PGPYGRXRYNPNMAPLVDV 1038
>BE319816 similar to PIR|G96707|G96 hypothetical protein T2E12.6 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 535
Score = 112 bits (279), Expect = 3e-25
Identities = 54/114 (47%), Positives = 83/114 (72%), Gaps = 2/114 (1%)
Frame = +3
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKA-KPVHV-SRYFVNRDIRSDQL 154
PK+AFMFLT G++ LW+ FF+GHE +S+YVH++ + H S F R I S ++
Sbjct: 159 PKVAFMFLTRGAVFLAPLWEQFFKGHEVYYSIYVHSNPSYNGSHPESPVFHGRRIPSKEV 338
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSF 208
WG ++++EAERRLL+NAL D +NQ FVL+S+SC+PL+NF+ I+ YL+ + +++
Sbjct: 339 EWGNVNMIEAERRLLSNALLDISNQRFVLISESCIPLFNFSTIYSYLINSTQNY 500
>TC80449 weakly similar to GP|15293267|gb|AAK93744.1 unknown protein
{Arabidopsis thaliana}, partial (56%)
Length = 1239
Score = 110 bits (274), Expect = 1e-24
Identities = 71/199 (35%), Positives = 103/199 (51%), Gaps = 19/199 (9%)
Frame = +3
Query: 126 FSVYVHASKAKPV-HVSRYFVNRDIRSDQLVWGKMSIVEAERRLLANAL-QDPNNQHFVL 183
F++Y+HA V F NR I S S++ A RRLLA+AL DP NQ+F L
Sbjct: 168 FNIYIHADPTTSVVSPGGVFHNRFISSKPTQRASPSLISAARRLLASALLDDPLNQYFAL 347
Query: 184 LSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD-----PGPVG------------NGRYSEH 226
+S CVPL +F ++++YL + SF D P + N R
Sbjct: 348 VSQHCVPLLSFRFVYNYLFKNQLMSLASFSDFNLLYPSFIEILSEDPNLYERYNARGENV 527
Query: 227 MLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPT 286
MLPEV +DFR G+Q+F L R+HA V+ D+ + KF+ CV+ +C +EHY PT
Sbjct: 528 MLPEVPFEDFRVGSQFFILNRKHAKVVVRDYKLWKKFRI----PCVNLDSCYPEEHYFPT 695
Query: 287 FFTIVDPNGIAKWSVTYVD 305
++ D NG +++T V+
Sbjct: 696 LLSMEDLNGCTGFTLTRVN 752
Score = 41.2 bits (95), Expect = 6e-04
Identities = 23/53 (43%), Positives = 32/53 (59%)
Frame = +2
Query: 90 TLDMPQNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSR 142
TL P+ PKIAF+FLT +L F LW+ FF G++ Y+H +P H+ R
Sbjct: 62 TLTNPK-PKIAFLFLTNTNLTFAPLWEKFFTGNQPSLQ-YIH--PCRPHHLRR 208
>TC82650 similar to GP|13374866|emb|CAC34500. putative protein {Arabidopsis
thaliana}, partial (41%)
Length = 784
Score = 82.4 bits (202), Expect = 2e-16
Identities = 63/184 (34%), Positives = 95/184 (51%), Gaps = 33/184 (17%)
Frame = +3
Query: 95 QNP--KIAFMFLTPGSLPFEKLWDNFFQGHEGK-FSVYVHASKAKPVHVSRYFVNRD--- 148
+NP KIAF+FLT L F LW+ FFQ K F+VYVH+ + + R N +
Sbjct: 213 KNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIF 392
Query: 149 --IRSDQLVWGKMSIVEAERRLLANA-LQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTD 205
I S + +++ A RRLLA+A L D +N +F++LS C+PL++F+YI+ L +
Sbjct: 393 KFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSP 572
Query: 206 ------------------KSFVDSFRDPGP------VGNGRYSEHMLPEVEIKDFRTGAQ 241
KSF++ + GP GRY+ M+PE + FR G+Q
Sbjct: 573 TFDLTDSESTQFGVRLKYKSFIEIINN-GPRLWKRYTARGRYA--MMPESPFEKFRVGSQ 743
Query: 242 WFSL 245
+F+L
Sbjct: 744 FFTL 755
>BG587356 weakly similar to GP|22535532|db hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 At5g14550, partial
(45%)
Length = 846
Score = 81.3 bits (199), Expect = 5e-16
Identities = 47/112 (41%), Positives = 67/112 (58%), Gaps = 15/112 (13%)
Frame = +3
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQ-GHEGKFSVYVHASKA----KPVHVSRYFVNRDI-R 150
PKIAF+FL ++P + LW FFQ G FS+YVH++ + S +F R +
Sbjct: 237 PKIAFLFLVRQNIPLDFLWGAFFQNGDVSNFSIYVHSTPGFLLDESTTRSHFFYGRQLSN 416
Query: 151 SDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDS---------CVPLYN 193
S Q++WG+ S+++AER LL+ AL+DP NQ FVLLSD C+ LY+
Sbjct: 417 SIQVLWGESSMIQAERLLLSAALEDPANQRFVLLSDRLCSSVQLFLCIQLYH 572
Score = 65.1 bits (157), Expect = 4e-11
Identities = 29/61 (47%), Positives = 43/61 (69%)
Frame = +1
Query: 188 CVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKR 247
CVPLYNF+Y+++Y+M + KSFVDSF D GRY+ M P++ + +R G+QW +L R
Sbjct: 529 CVPLYNFSYVYNYIMVSPKSFVDSFLD---AKEGRYNPKMSPKIPREKWRKGSQWVTLVR 699
Query: 248 Q 248
+
Sbjct: 700 K 702
>BF649927 similar to GP|17065506|gb| Unknown protein {Arabidopsis thaliana},
partial (8%)
Length = 633
Score = 65.9 bits (159), Expect = 2e-11
Identities = 28/33 (84%), Positives = 30/33 (90%)
Frame = +2
Query: 98 KIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYV 130
K+ FMFLTPG+LPFEKLW FFQGHEGKFSVYV
Sbjct: 533 KLLFMFLTPGTLPFEKLWHTFFQGHEGKFSVYV 631
Score = 48.1 bits (113), Expect = 5e-06
Identities = 25/70 (35%), Positives = 41/70 (57%), Gaps = 1/70 (1%)
Frame = +1
Query: 1 MKTAKFWRLSMGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTS-C 59
MK W ++ I++GS HR +K+P WI+VLV + FL AY+Y +++ S C
Sbjct: 199 MKKEFAWPQFTKNLLIMVGSR--HRSNLKRPTWIIVLVFIVCFFLLAAYIYPPRSSASTC 372
Query: 60 NMFYSKPCII 69
++F S+ C +
Sbjct: 373 SLFNSQGCSV 402
>BE943485 similar to PIR|T48532|T485 hypothetical protein T22P22.120 -
Arabidopsis thaliana, partial (20%)
Length = 487
Score = 54.3 bits (129), Expect = 7e-08
Identities = 34/106 (32%), Positives = 50/106 (47%)
Frame = +1
Query: 252 KVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKR 311
K+ + + Y+ F EQ C + C +DEHY PT TI + +A SVT+VD S
Sbjct: 7 KLAINIVVYTAFYP*WEQYC--RRACYVDEHYFPTMLTIQAGSALANRSVTWVDWSRGGA 180
Query: 312 HPKSYRTQDITYELLKNIKLHNFHLFLTERSTKMDLFLEWISEAVL 357
HP ++ DIT E ++ L+ ST LF + + L
Sbjct: 181 HPATFGRTDITNEFFDRVRGGQKCLYNNRNSTLCSLFARKFAPSAL 318
>TC83971 weakly similar to GP|23315108|gb|AAN20289.1 Sequence 273 from
patent US 6410718, partial (41%)
Length = 683
Score = 50.1 bits (118), Expect = 1e-06
Identities = 18/37 (48%), Positives = 28/37 (75%)
Frame = +2
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHAS 133
PK+AF+FL G +P LW+ FF+GH+G +S+ VH++
Sbjct: 569 PKVAFLFLVRGPVPLAPLWEKFFKGHKGYYSIXVHSN 679
>BE998206 weakly similar to GP|20804766|dbj contains ESTs AU096118(S12194)
AU096119(S12194)~unknown protein, partial (13%)
Length = 446
Score = 39.3 bits (90), Expect = 0.002
Identities = 18/55 (32%), Positives = 29/55 (52%)
Frame = -2
Query: 276 NCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIK 330
+C DEHYLPT +I G + S+ +VD S HP + D+ + L+ ++
Sbjct: 442 SCYADEHYLPTLVSIKFGEGNSNRSLPWVDWSRGGPHPAHFLRSDVNVKFLERLR 278
>BF521022 homologue to GP|17944908|gb LP04958p {Drosophila melanogaster},
partial (90%)
Length = 570
Score = 29.6 bits (65), Expect = 1.9
Identities = 19/61 (31%), Positives = 27/61 (44%)
Frame = -1
Query: 239 GAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAK 298
G Q +RQH + HL + Q QSC D C+L L F I+ G+++
Sbjct: 429 GIQHHQYQRQHHPSCLC-HLVHVCMQNDLRQSCFDHHRCVL*LLVLDALFLIL--LGVSR 259
Query: 299 W 299
W
Sbjct: 258 W 256
>AL366786 similar to PIR|C86405|C86 probable sphingosine-1-phosphate lyase
[imported] - Arabidopsis thaliana, partial (20%)
Length = 505
Score = 29.6 bits (65), Expect = 1.9
Identities = 18/51 (35%), Positives = 25/51 (48%)
Frame = +1
Query: 171 NALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNG 221
NALQ PN+ H CV L + I D+L ++S +PGP+ G
Sbjct: 121 NALQRPNSIHI------CVTLQHVPVIEDFLRDLNESVKTVKENPGPITGG 255
>BQ144447 GP|12697999|d KIAA1727 protein {Homo sapiens}, partial (1%)
Length = 1379
Score = 28.1 bits (61), Expect = 5.5
Identities = 17/67 (25%), Positives = 33/67 (48%)
Frame = -2
Query: 17 LLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSCNMFYSKPCIIDISVLAV 76
+L +L P + +P+ + FII F T AY+ + + ++ + + + +S L
Sbjct: 709 MLSLSLLPNPPLSRPLLFSHIFLFIIYFYT-AYICSISSVSAPLLLLLQALLHSLSCLRP 533
Query: 77 SHIPVSV 83
HIP +V
Sbjct: 532 LHIPTAV 512
>TC89451 weakly similar to GP|21536959|gb|AAM61300.1 unknown {Arabidopsis
thaliana}, partial (80%)
Length = 1609
Score = 27.7 bits (60), Expect = 7.1
Identities = 10/24 (41%), Positives = 14/24 (57%)
Frame = -2
Query: 24 HRPQMKKPMWIVVLVSFIILFLTC 47
H + +P W V V F+I+F TC
Sbjct: 1101 HNSKRIRPYWFVHAVGFLIMFATC 1030
>BF645824 similar to PIR|T07142|T07 beta-glucan-elicitor receptor - soybean,
partial (18%)
Length = 629
Score = 27.7 bits (60), Expect = 7.1
Identities = 41/163 (25%), Positives = 70/163 (42%), Gaps = 4/163 (2%)
Frame = +3
Query: 38 VSFIILFLTCAYLYRMQNTTSCNMFYSKPCIIDISVLAVSHIP----VSVYIERGITLDM 93
VS+ +L + A LY++ +S I S ++IP +S Y + G+TLD+
Sbjct: 189 VSYPLLLFSTAMLYQV---------FSPDLTISSSQKTHTNIPKNHVISSYSDLGVTLDI 341
Query: 94 PQNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQ 153
P + + F FL GS PF + AS KP+ +S ++ I
Sbjct: 342 PSS-NLRF-FLVRGS-PF------------------LTASVTKPIPLSITTIHSII---- 446
Query: 154 LVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNY 196
S+ +++ LQ NNQ +++ + S + NFN+
Sbjct: 447 ------SLSPFDKKKTKYTLQLNNNQTWIIYTSSPI---NFNH 548
>TC83191 similar to GP|6714272|gb|AAF25968.1| F6N18.8 {Arabidopsis
thaliana}, partial (58%)
Length = 1053
Score = 27.3 bits (59), Expect = 9.3
Identities = 16/52 (30%), Positives = 28/52 (53%)
Frame = +3
Query: 11 MGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSCNMF 62
MGD E ++G L+ ++K P W+ L+ F+ C ++ N + CNM+
Sbjct: 93 MGDGEGVVGVRSLNEEEIKWPPWLQPLLQ-ARFFVQCK-VHADSNKSECNMY 242
>BF646214 similar to GP|8843779|dbj| gene_id:MZN1.3~unknown protein
{Arabidopsis thaliana}, partial (27%)
Length = 638
Score = 27.3 bits (59), Expect = 9.3
Identities = 14/46 (30%), Positives = 20/46 (43%)
Frame = +2
Query: 297 AKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIKLHNFHLFLTERS 342
AKW + Q R + RT ++L + H+FH F E S
Sbjct: 128 AKWFLAQAHAQAQARRRRRRRTTVTVSDVLGPARFHHFHTFNIEDS 265
>TC86469 similar to GP|21464654|emb|CAD30328. acid phosphatase {Lupinus
luteus}, partial (92%)
Length = 1737
Score = 27.3 bits (59), Expect = 9.3
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 7/40 (17%)
Frame = +3
Query: 247 RQHAVKVMADHLYYSKFQAQ-------CEQSCVDGKNCIL 279
R + +V++ +YYS F Q C Q+C+ NC++
Sbjct: 765 RSCSFQVLSSTIYYSLFGVQEYQPPLVCNQACICSYNCVI 884
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.139 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,981,801
Number of Sequences: 36976
Number of extensions: 201472
Number of successful extensions: 1472
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1455
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1461
length of query: 359
length of database: 9,014,727
effective HSP length: 97
effective length of query: 262
effective length of database: 5,428,055
effective search space: 1422150410
effective search space used: 1422150410
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149130.7


