

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.11 - phase: 0 /pseudo
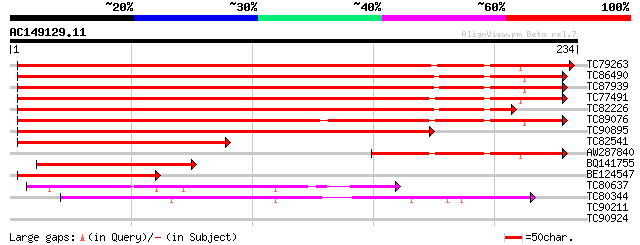
(234 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79263 similar to GP|6016736|gb|AAF01562.1| unknown protein {Ar... 295 1e-80
TC86490 similar to PIR|T48615|T48615 hypothetical protein F18O22... 272 7e-74
TC87939 similar to PIR|T48615|T48615 hypothetical protein F18O22... 267 2e-72
TC77491 similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20 ... 246 7e-66
TC82226 similar to PIR|T48615|T48615 hypothetical protein F18O22... 240 4e-64
TC89076 similar to GP|18481708|gb|AAL73530.1 hypothetical protei... 225 1e-59
TC90895 similar to GP|21689819|gb|AAM67553.1 unknown protein {Ar... 209 7e-55
TC82541 similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20 ... 104 3e-23
AW287840 similar to GP|13937149|gb At1g79380/T8K14_20 {Arabidops... 83 8e-17
BQ141755 similar to GP|18481708|gb| hypothetical protein {Sorghu... 80 5e-16
BE124547 similar to GP|18481708|gb hypothetical protein {Sorghum... 75 3e-14
TC80637 similar to GP|14488104|gb|AAK63871.1 AT5g07300/T2I1_10 {... 68 3e-12
TC80344 similar to GP|9758389|dbj|BAB08876.1 copine-like protein... 57 5e-09
TC90211 similar to SP|P93768|PSD3_TOBAC Probable 26S proteasome ... 28 4.0
TC90924 similar to GP|22758266|gb|AAN05494.1 Putative F-box prot... 27 5.2
>TC79263 similar to GP|6016736|gb|AAF01562.1| unknown protein {Arabidopsis
thaliana}, partial (58%)
Length = 1419
Score = 295 bits (754), Expect = 1e-80
Identities = 159/231 (68%), Positives = 178/231 (76%), Gaps = 1/231 (0%)
Frame = +3
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GK+S R+SLH I N PN E+AISIIGK + F +DNLIPCFGFGDAST D DVFSFY
Sbjct: 540 TGKHSFNRKSLHHIGNGPNPYEQAISIIGKTLAAFDEDNLIPCFGFGDASTHDQDVFSFY 719
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
DER CNG EE+LSRYREI PNI+ AG TSFAPI+E A IV+QSGG++HVLVII+DGQV
Sbjct: 720 PDERICNGFEEVLSRYREIVPNIKLAGPTSFAPIVETAMTIVEQSGGQYHVLVIIADGQV 899
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D +HG LS Q+ TVDAIVEA KFPLSIILVGVGDGPWDM+KEF DN M R F
Sbjct: 900 TRSIDTEHGRLSPQEQKTVDAIVEASKFPLSIILVGVGDGPWDMMKEFDDN--MPARAFD 1073
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEI-LLAIREILFQYKAAKELGLLGLTK 233
NF+FVNFTE+MSK NI PS KEA L A+ EI QYKAA EL LLG K
Sbjct: 1074NFQFVNFTELMSK--NIPPSRKEAAFALAALMEIPSQYKAAIELNLLGSRK 1220
>TC86490 similar to PIR|T48615|T48615 hypothetical protein F18O22.210 -
Arabidopsis thaliana, partial (72%)
Length = 1739
Score = 272 bits (696), Expect = 7e-74
Identities = 143/228 (62%), Positives = 177/228 (76%), Gaps = 1/228 (0%)
Frame = +3
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GK S R+SLH I + N E+AISIIGK S F +DNLIPCFGFGDAST D DVFSFY
Sbjct: 435 TGKSSFNRKSLHHIGSGQNPYEQAISIIGKTLSTFDEDNLIPCFGFGDASTHDQDVFSFY 614
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
+ER CNG EE+L+RYREI P ++ AG TSFAPIIEMA IV+QS G++HVL+II+DGQV
Sbjct: 615 SEERLCNGFEEVLARYREIVPQLKLAGPTSFAPIIEMAMTIVEQSAGQYHVLLIIADGQV 794
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D ++G+LS Q+ T++AIV+A ++PLSI+LVGVGDGPW+M+KEF DNI +R F
Sbjct: 795 TRSIDTEYGNLSPQEQKTINAIVKASEYPLSIVLVGVGDGPWEMMKEFDDNI--PSRAFD 968
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFT+IMSK N++PS +E E L A+ EI QYKA ELGLLG
Sbjct: 969 NFQFVNFTKIMSK--NVNPSRRETEFALDALMEIPSQYKATIELGLLG 1106
>TC87939 similar to PIR|T48615|T48615 hypothetical protein F18O22.210 -
Arabidopsis thaliana, partial (69%)
Length = 1612
Score = 267 bits (683), Expect = 2e-72
Identities = 142/228 (62%), Positives = 173/228 (75%), Gaps = 1/228 (0%)
Frame = +1
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G S R+ LHDI + PN E+AISIIGK S F +DNLIPCFGFGDAST D +VFSF
Sbjct: 436 TGARSFQRRCLHDIGHEPNPYEQAISIIGKTLSSFDEDNLIPCFGFGDASTHDQEVFSFN 615
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
++RFC+G EE+LSRYRE+ P ++ AG TSFAPIIEMA IV+QS G++HVL+II+DGQV
Sbjct: 616 AEDRFCHGFEEVLSRYRELVPQLRLAGPTSFAPIIEMAITIVEQSEGQYHVLLIIADGQV 795
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D HG LS+Q+ TV+AIV+A ++PLSI+LVGVGDGPWDM+K+F DNI R F
Sbjct: 796 TRSVDVDHGQLSAQEKKTVEAIVKASEYPLSIVLVGVGDGPWDMMKQFDDNI--PARAFD 969
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFTEIMSK N+ S KEAE L A+ EI QYKA EL +LG
Sbjct: 970 NFQFVNFTEIMSK--NMDRSRKEAEFALSALMEIPDQYKATLELSILG 1107
>TC77491 similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20
{Arabidopsis thaliana}, partial (81%)
Length = 1537
Score = 246 bits (627), Expect = 7e-66
Identities = 129/228 (56%), Positives = 166/228 (72%), Gaps = 1/228 (0%)
Frame = +2
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G+ S +SLH I PN EKAISIIGK +PF DDNLIPC GFGDA+T D +VFSF+
Sbjct: 323 TGRSSFNNKSLHSIGGTPNPYEKAISIIGKTLAPFDDDNLIPCLGFGDATTHDSEVFSFH 502
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
D+ C+G EE+L+ Y++I PN+ +G TS+AP+IE A IV+++ G+ HVLVI++DGQV
Sbjct: 503 ADDSSCHGFEEVLACYQKIVPNLSLSGPTSYAPVIEAAMDIVEKTHGQFHVLVIVADGQV 682
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R + + G LS Q+ T+ AI +A +PLSI+LVGVGDGPW+ +K+F D K+ R F
Sbjct: 683 TRYGNNEGGKLSPQEEKTIKAIADASSYPLSIVLVGVGDGPWEDMKKFDD--KLPARDFD 856
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEI-LLAIREILFQYKAAKELGLLG 230
NF+FVNFT+IMSK ISPS KEA L A+ EI FQYKAA ELGLLG
Sbjct: 857 NFQFVNFTDIMSK--KISPSAKEAAFALAALMEIPFQYKAALELGLLG 994
>TC82226 similar to PIR|T48615|T48615 hypothetical protein F18O22.210 -
Arabidopsis thaliana, partial (63%)
Length = 1300
Score = 240 bits (612), Expect = 4e-64
Identities = 123/206 (59%), Positives = 155/206 (74%)
Frame = +2
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GK S R+SLH I N N E+AI+IIGK + F +DN IPCFGFGDAST D DVFSF+
Sbjct: 125 TGKKSFNRKSLHHIGNGQNPYEQAIAIIGKTLTAFDEDNSIPCFGFGDASTHDQDVFSFH 304
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
DER CNG EE+LS+Y EI P ++ A TSFAPIIEM IV++SGG++HV +II+DGQV
Sbjct: 305 SDERCCNGFEEVLSKYTEIVPCLRLARPTSFAPIIEMGMTIVEKSGGQYHVSLIIADGQV 484
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
R D ++G LSSQ+ T+DAIV+A ++PLSI+LVGVGDGPWDM++EF DNI R F
Sbjct: 485 TRSVDTENGKLSSQEKKTIDAIVKASEYPLSIVLVGVGDGPWDMMREFDDNI--PARAFD 658
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEI 209
NF+FVNFTEIMS+ N+ + KE +
Sbjct: 659 NFQFVNFTEIMSR--NVDSNRKETRV 730
>TC89076 similar to GP|18481708|gb|AAL73530.1 hypothetical protein {Sorghum
bicolor}, partial (65%)
Length = 1326
Score = 225 bits (574), Expect = 1e-59
Identities = 121/228 (53%), Positives = 164/228 (71%), Gaps = 1/228 (0%)
Frame = +2
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G+ S ++SLH I + PN EKAISI+GK +PF +DNLIPCFGFGDA+T D +VFSF+
Sbjct: 272 TGQISFNKKSLHAIGDTPNPYEKAISIVGKTLAPFDEDNLIPCFGFGDATTHDQEVFSFH 451
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
D C+G EE+L+ Y+ I PN++ +G TS+AP+IE A IV++S G+ HVLVII+DGQV
Sbjct: 452 SDHSPCHGFEEVLACYKNIVPNLKLSGPTSYAPVIEAAIDIVEKSHGQFHVLVIIADGQV 631
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFH 183
+ D +S Q+ T+ AI +A K+PL+I+LVGVGDGPW+ +++F D K+ +R F
Sbjct: 632 TKGVD---NEVSPQEKKTIKAIADASKYPLAIVLVGVGDGPWEDMEKFDD--KIPSRDFD 796
Query: 184 NFKFVNFTEIMSKSNNISPSLKEAEILL-AIREILFQYKAAKELGLLG 230
NF+FVNFT+IMSK N S + KEA + A+ EI FQYKA EL LG
Sbjct: 797 NFQFVNFTKIMSK--NTSAAEKEAAFAVNALMEIPFQYKACVELQKLG 934
>TC90895 similar to GP|21689819|gb|AAM67553.1 unknown protein {Arabidopsis
thaliana}, partial (58%)
Length = 1018
Score = 209 bits (532), Expect = 7e-55
Identities = 101/172 (58%), Positives = 130/172 (74%)
Frame = +1
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+GKYS R+SLH I + PN E+AISIIG+ S F +DNLIPCFGFGDAST D +VFSFY
Sbjct: 493 TGKYSFHRKSLHHIGSAPNPYEQAISIIGRTLSTFDEDNLIPCFGFGDASTHDQNVFSFY 672
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQSGGKHHVLVIISDGQV 123
D R C+G EE+L+RYR+I P+++ +G TSFAP+I+ A IV+++ G++HVLVII+DGQV
Sbjct: 673 PDNRVCHGFEEVLARYRQIVPHLKLSGPTSFAPVIDAAIDIVERNNGQYHVLVIIADGQV 852
Query: 124 IRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVGDGPWDMVKEFQDNI 175
R HG LS Q+ T+ +I+ A +PLSIILVGVGDGPWD +K DNI
Sbjct: 853 TRNPGTPHGKLSPQEQATITSIIAASHYPLSIILVGVGDGPWDEMKHXDDNI 1008
>TC82541 similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20
{Arabidopsis thaliana}, partial (32%)
Length = 820
Score = 104 bits (259), Expect = 3e-23
Identities = 48/88 (54%), Positives = 66/88 (74%)
Frame = +1
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFY 63
+G+ S ++SLH I + PN EKAISI+GK +PF +DNLIPCFGFGDA+T D +VFSF+
Sbjct: 514 TGQVSFNKRSLHAIGDTPNPYEKAISIVGKTLAPFDEDNLIPCFGFGDATTHDQEVFSFH 693
Query: 64 RDERFCNGHEEILSRYREIRPNIQPAGT 91
D C+G EE+L+ Y+ I PN++ +GT
Sbjct: 694 SDHSPCHGFEEVLACYKNIVPNLKLSGT 777
>AW287840 similar to GP|13937149|gb At1g79380/T8K14_20 {Arabidopsis
thaliana}, partial (33%)
Length = 475
Score = 83.2 bits (204), Expect = 8e-17
Identities = 46/82 (56%), Positives = 59/82 (71%), Gaps = 1/82 (1%)
Frame = +2
Query: 150 KFPLSIILVGVGDGPWDMVKEFQDNIKMQTRTFHNFKFVNFTEIMSKSNNISPSLKEAEI 209
K+PL+I+L+GVGDGPW+ +++F D K+ TR F NF+FVNFT+IMSK N S + KEA
Sbjct: 2 KYPLAIVLIGVGDGPWEDMEKFDD--KLPTRDFDNFQFVNFTKIMSK--NTSETEKEAAF 169
Query: 210 -LLAIREILFQYKAAKELGLLG 230
L A+ EI FQYKA E LG
Sbjct: 170 ALAALMEIPFQYKACVEFKKLG 235
>BQ141755 similar to GP|18481708|gb| hypothetical protein {Sorghum bicolor},
partial (12%)
Length = 702
Score = 80.5 bits (197), Expect = 5e-16
Identities = 37/66 (56%), Positives = 48/66 (72%)
Frame = +3
Query: 12 QSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFYRDERFCNG 71
++L I PN EKAISIIGK +PF DDNLIPC G+GDA+T D + FSF+ D+ C+G
Sbjct: 312 ENLRCIGGPPNPYEKAISIIGKTLAPFDDDNLIPCLGYGDATTHDTETFSFHADDSSCHG 491
Query: 72 HEEILS 77
EE+L+
Sbjct: 492 FEEVLA 509
>BE124547 similar to GP|18481708|gb hypothetical protein {Sorghum bicolor},
partial (22%)
Length = 525
Score = 74.7 bits (182), Expect = 3e-14
Identities = 35/59 (59%), Positives = 45/59 (75%)
Frame = +3
Query: 4 SGKYSLIRQSLHDIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSF 62
+G+ S ++SLH I + N EKAISI+GK +PF +DNLIPCFGFGDA+T D +VFSF
Sbjct: 348 TGQISFNKKSLHAIGDTLNPYEKAISIVGKTLAPFDEDNLIPCFGFGDATTHDQEVFSF 524
>TC80637 similar to GP|14488104|gb|AAK63871.1 AT5g07300/T2I1_10 {Arabidopsis
thaliana}, partial (50%)
Length = 1302
Score = 68.2 bits (165), Expect = 3e-12
Identities = 59/165 (35%), Positives = 79/165 (47%), Gaps = 11/165 (6%)
Frame = +1
Query: 8 SLIRQSLH--DIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDV---FSF 62
S + SLH D PNS ++AI +G+ + D P +GFG A D V F+
Sbjct: 226 SRLPDSLHYIDPSGRPNSYQRAIIEVGEVLQFYDSDKRFPTWGFG-ARPIDGPVSHCFNL 402
Query: 63 YRDERFCN--GHEEILSRYREIRPNIQPAGTTSFAPIIEMATKIVDQS----GGKHHVLV 116
+C G + I+ Y N+ AG T F P+I A I QS G K+ VL+
Sbjct: 403 NGSNTYCEVEGIQGIMMAYTSALLNVSLAGPTLFGPVINNAAIIASQSVANGGRKYFVLL 582
Query: 117 IISDGQVIRRNDAQHGSLSSQKLHTVDAIVEARKFPLSIILVGVG 161
II+DG V D Q T DA+V+A PLSI++VGVG
Sbjct: 583 IITDGVV---TDLQE---------TKDALVKASDLPLSILIVGVG 681
>TC80344 similar to GP|9758389|dbj|BAB08876.1 copine-like protein
{Arabidopsis thaliana}, partial (35%)
Length = 887
Score = 57.4 bits (137), Expect = 5e-09
Identities = 55/218 (25%), Positives = 95/218 (43%), Gaps = 22/218 (10%)
Frame = +2
Query: 22 NSCEKAISIIGKKFSPFLDDNLIPCFGFGDASTSDHDVFSFYRD-----ERFCNGHEEIL 76
NS +K I +G+ + D L P +GFG F + G E I+
Sbjct: 170 NSYQKVIMEVGEVIQFYDSDKLFPAWGFGGNVHGGSVSHCFNLNGGAPGSSEVEGIEGIM 349
Query: 77 SRYREIRPNIQPAGTTSFAPIIEMATKIVDQS-----GGKHHVLVIISDGQVIRRNDAQH 131
Y ++ +G T F P+I +A ++ S K++VL+II+DG V +++
Sbjct: 350 DAYGSALRSVTLSGPTLFGPVINLAAQLAADSLSSYNSSKYYVLLIITDGIVTDLHES-- 523
Query: 132 GSLSSQKLHTVDAIVEARKFPLSIILVGVGDGP-----WDMVKEFQDNIKMQT---RTFH 183
++A+V A PLSI++VGVG P W ++ + + + +
Sbjct: 524 ----------INAVVNASDLPLSILIVGVGLIPPAWRYWILIMDLA*KVLLVV*LHEI*Y 673
Query: 184 NF----KFVNFTEIMSKSNNISPSLKEAEILLAIREIL 217
N K+ T + S+N+ L I+L++R+IL
Sbjct: 674 NLSP*EKYKVSTIVACFSSNLFNGLSVLRIILSLRDIL 787
>TC90211 similar to SP|P93768|PSD3_TOBAC Probable 26S proteasome non-ATPase
regulatory subunit 3 (26S proteasome subunit S3),
partial (57%)
Length = 1009
Score = 27.7 bits (60), Expect = 4.0
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = +2
Query: 14 LHDIRNVPNSCEKAISIIGKKFSPF 38
LHD+R++ S AIS KF PF
Sbjct: 734 LHDLRHIQTSSSVAISSTSGKFGPF 808
>TC90924 similar to GP|22758266|gb|AAN05494.1 Putative F-box protein
{Oryza sativa (japonica cultivar-group)}, partial (15%)
Length = 819
Score = 27.3 bits (59), Expect = 5.2
Identities = 13/38 (34%), Positives = 19/38 (49%)
Frame = +1
Query: 16 DIRNVPNSCEKAISIIGKKFSPFLDDNLIPCFGFGDAS 53
D+ N+PN + + + K PFLD + C GD S
Sbjct: 31 DLSNLPNLSDNGVLELAKSPIPFLDLRMRQCPLIGDTS 144
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,743,920
Number of Sequences: 36976
Number of extensions: 84977
Number of successful extensions: 401
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 387
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 388
length of query: 234
length of database: 9,014,727
effective HSP length: 93
effective length of query: 141
effective length of database: 5,575,959
effective search space: 786210219
effective search space used: 786210219
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149129.11


