

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149127.4 + phase: 0
(323 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
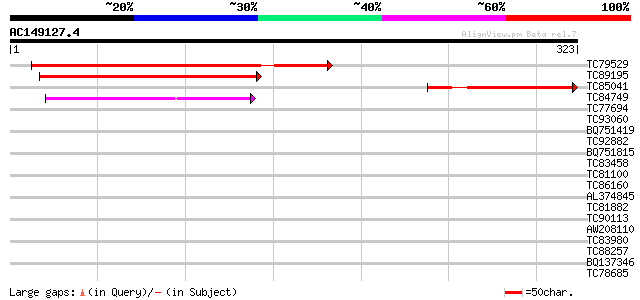
Score E
Sequences producing significant alignments: (bits) Value
TC79529 similar to GP|20197375|gb|AAM15048.1 expressed protein {... 143 1e-34
TC89195 similar to GP|20197375|gb|AAM15048.1 expressed protein {... 133 1e-31
TC85041 similar to PIR|T45834|T45834 hypothetical protein F2K15.... 116 1e-26
TC84749 weakly similar to GP|14587302|dbj|BAB61213. hypothetical... 73 2e-13
TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-... 39 0.002
TC93060 weakly similar to PIR|S55970|S55970 ribosomal protein L6... 38 0.006
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 37 0.013
TC92882 similar to PIR|T43257|T43257 beta-1 3 exoglucanase (EC 3... 37 0.013
BQ751815 similar to PIR|S58652|S586 hypothetical protein YFR036w... 36 0.018
TC83458 homologue to PIR|T07796|T07796 DNA-directed RNA polymera... 35 0.030
TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon prot... 35 0.051
TC86160 weakly similar to PIR|T01605|T01605 phytocyanin At2g4479... 34 0.067
AL374845 similar to GP|12247468|gb| PnFL-1 {Ipomoea nil}, partia... 33 0.11
TC81882 similar to GP|9759349|dbj|BAB10004.1 contains similarity... 33 0.15
TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protei... 33 0.19
AW208110 similar to GP|14715462|d CjMDR1 {Coptis japonica}, part... 32 0.33
TC83980 similar to GP|10177915|dbj|BAB11326. gene_id:MCA23.11~un... 32 0.43
TC88257 weakly similar to PIR|G96668|G96668 protein F1N19.7 [imp... 31 0.56
BQ137346 homologue to GP|15027095|em hypothetical protein LT.09 ... 31 0.74
TC78685 weakly similar to GP|9757770|dbj|BAB08379.1 gene_id:MOK1... 30 0.96
>TC79529 similar to GP|20197375|gb|AAM15048.1 expressed protein {Arabidopsis
thaliana}, partial (74%)
Length = 1378
Score = 143 bits (360), Expect = 1e-34
Identities = 75/172 (43%), Positives = 108/172 (62%)
Frame = +3
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
A +DEV M ++ F ++L +LKNLR QLYSAAEY E SY + +QKQ++++ LKDYAV+A
Sbjct: 171 AMTFDEVSMERSKSFVNALQELKNLRPQLYSAAEYCEKSYLHSEQKQMVLDNLKDYAVRA 350
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
L+N VDHLG+VAYK++DLL++ +V L++S I Q++ TCQ + D EG QQ L+
Sbjct: 351 LVNAVDHLGTVAYKLTDLLEQHTLDVSTMGLKVSTINQKLHTCQIYTDKEGLRQQQLLAF 530
Query: 133 TPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETP 184
P+HHK YILP + K + H+ + + F+ R TP
Sbjct: 531 IPRHHKHYILP-------NSVNKKVHFSPHIQIDAKQNSFQTRTRFQSSGTP 665
>TC89195 similar to GP|20197375|gb|AAM15048.1 expressed protein {Arabidopsis
thaliana}, partial (55%)
Length = 896
Score = 133 bits (334), Expect = 1e-31
Identities = 65/126 (51%), Positives = 90/126 (70%)
Frame = +3
Query: 18 EVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKALINTV 77
E M Q+ F +L +LKNLR QLYSAA+Y E SY DQK+I+++ +KDYA +AL+N V
Sbjct: 93 EFAMEQSKSFVFALQELKNLRPQLYSAADYCEKSYLRSDQKEIVLDNMKDYAARALVNAV 272
Query: 78 DHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVISTPKHH 137
DHLG+VA K+SDLL+++ +V DL++S + QR+ TCQ + + EG QQ L+ P+HH
Sbjct: 273 DHLGTVACKLSDLLEQQTLDVSTMDLKISTLNQRLLTCQLYTNKEGVRQQQLLAIIPRHH 452
Query: 138 KRYILP 143
K YILP
Sbjct: 453 KHYILP 470
>TC85041 similar to PIR|T45834|T45834 hypothetical protein F2K15.150 -
Arabidopsis thaliana, partial (19%)
Length = 464
Score = 116 bits (291), Expect = 1e-26
Identities = 60/85 (70%), Positives = 68/85 (79%)
Frame = +1
Query: 239 RSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSK 298
RSTTPK R TPN PSN + RYPSEPRKSASMRLS++ +N +D++Q PSKSK
Sbjct: 1 RSTTPKPSRPITPN--------PSNTKQRYPSEPRKSASMRLSAERDNGKDVEQFPSKSK 156
Query: 299 RLLKSLLSRRKSKKDDTLYTYLDEY 323
RLLK+LLSRRKSKKDDTLYTYLDEY
Sbjct: 157 RLLKALLSRRKSKKDDTLYTYLDEY 231
>TC84749 weakly similar to GP|14587302|dbj|BAB61213. hypothetical protein
{Oryza sativa (japonica cultivar-group)}, partial (39%)
Length = 650
Score = 72.8 bits (177), Expect = 2e-13
Identities = 35/120 (29%), Positives = 67/120 (55%)
Frame = +1
Query: 21 MHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKALINTVDHL 80
+ + + F SL +L+ LR+QL+ AA+Y E ++ ++K+ +++ K+Y +A++ VDHL
Sbjct: 160 VEENMHFLKSLQELRELRSQLHYAADYCETTFMESEKKRDVMDDTKEYICRAMVTVVDHL 339
Query: 81 GSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVISTPKHHKRY 140
G+V+ + L+ K ++R+ C+ QR+ TC + D + P+ H RY
Sbjct: 340 GNVSSNLEGLISHK-NSFSDAEIRIQCLTQRLFTCDQYADKVALSNMQWREKLPRLHTRY 516
>TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-rich
splicing factor - alfalfa, partial (95%)
Length = 1494
Score = 39.3 bits (90), Expect = 0.002
Identities = 32/115 (27%), Positives = 48/115 (40%), Gaps = 3/115 (2%)
Frame = +3
Query: 151 TNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSS---KGNSPSPSLQPQRVGAFS 207
+ S Y G DD R+ R+ +P SS + SP S P+R
Sbjct: 534 SRSASPGYKGRGRGRHDDERRSRSPSRSVDSRSPVRRSSIPKRSPSPKRSPSPKR----- 698
Query: 208 FTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPS 262
SP++ + +++VSP + PL + SR T RS +P A+ SPS
Sbjct: 699 --SPSLKRSISPQKSVSPRKSPLRESPDNRSRGGRSLTPRSVSPRGRPGASRSPS 857
>TC93060 weakly similar to PIR|S55970|S55970 ribosomal protein L6.e.B
cytosolic - yeast (Saccharomyces cerevisiae), partial
(77%)
Length = 903
Score = 37.7 bits (86), Expect = 0.006
Identities = 33/137 (24%), Positives = 58/137 (42%), Gaps = 1/137 (0%)
Frame = +3
Query: 177 RATIRETPTSTSSKGNSPSPS-LQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGS 235
R + R + T + +SP+PS A S +S + + +P R S +
Sbjct: 108 RRSARPSAPGTLASPSSPAPS*FSLPVASAASASSS*RPSTRVSSSSPAPSRSTASPSAV 287
Query: 236 MSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPS 295
+ +++P RST+P S+ R N P+ PR + RLSS ++ P
Sbjct: 288 STPATSSPHPTRSTSPLSTRRRLRRSRNPSTSPPTRPRPRPARRLSS------SRERSPR 449
Query: 296 KSKRLLKSLLSRRKSKK 312
+ + +L +RR S +
Sbjct: 450 RRRSTAAALPTRRPSTR 500
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 36.6 bits (83), Expect = 0.013
Identities = 30/101 (29%), Positives = 47/101 (45%), Gaps = 3/101 (2%)
Frame = +1
Query: 185 TSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPK 244
T++S+ +P+PS P R + +P A + +P P + S S+ +TT
Sbjct: 94 TASSTPPGAPTPSPTPTRRATLNGNTPAPASPTAQ---ATPPTGPSTAAPSPSTCTTTGP 264
Query: 245 TGRSTT---PNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
T ST+ P +++ TTSPS P P SAS +S
Sbjct: 265 TSSSTSASSPAATSPRTTSPSPHSSGTPPAPAPSASRSSTS 387
>TC92882 similar to PIR|T43257|T43257 beta-1 3 exoglucanase (EC 3.2.1.-)
precursor - fungus (Trichoderma harzianum), partial (3%)
Length = 627
Score = 36.6 bits (83), Expect = 0.013
Identities = 32/99 (32%), Positives = 43/99 (43%), Gaps = 4/99 (4%)
Frame = -1
Query: 181 RETPTSTSSKGNSPSPSLQ--PQRVG--AFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSM 236
R +PTSTS+ P PS + R G A S TSP+ A S R + + +
Sbjct: 591 RPSPTSTSTSPRDPVPSTRASSSRKGPAACSTTSPSTAGCTAPTSATSSTRSATAASSAA 412
Query: 237 SSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKS 275
S ST+ TG T S++R S +R P R S
Sbjct: 411 RSPSTSSGTGAGRTRASTSRTAPWASGSRTTAPRR*RCS 295
>BQ751815 similar to PIR|S58652|S586 hypothetical protein YFR036w-a - yeast
(Saccharomyces cerevisiae), partial (9%)
Length = 719
Score = 36.2 bits (82), Expect = 0.018
Identities = 26/95 (27%), Positives = 42/95 (43%)
Frame = +1
Query: 182 ETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRST 241
E+PTS +GNS S P + A + + + + K S P + R+
Sbjct: 127 ESPTSHPRRGNSLWRSFSPSSLVASRTSFSSTTRPEPCKPPSSTRPPPKGNKSPANYRAP 306
Query: 242 TPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSA 276
TP + R+ TP+SS+ + + R R S P +A
Sbjct: 307 TPNSPRAGTPSSSSPSWSCRPTPRSRTSSSPSSTA 411
Score = 30.8 bits (68), Expect = 0.74
Identities = 34/115 (29%), Positives = 43/115 (36%), Gaps = 11/115 (9%)
Frame = +1
Query: 178 ATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMS 237
++ R P S N +P+ R G S +SP+ + + P SRT S
Sbjct: 250 SSTRPPPKGNKSPANYRAPTPNSPRAGTPSSSSPSWSCRPT----------PRSRTSSSP 399
Query: 238 SRSTTPKTGRSTTPNSSNRATTSPSNARVR-----------YPSEPRKSASMRLS 281
S ST STTP TTS R R PS P SA+ R S
Sbjct: 400 S-STARSAS*STTPRPRGFWTTSTDKLRPRSKSPFF*ENGTAPSLPSWSATRRRS 561
>TC83458 homologue to PIR|T07796|T07796 DNA-directed RNA polymerase (EC
2.7.7.6) largest chain - soybean (fragment), partial
(63%)
Length = 1220
Score = 35.4 bits (80), Expect = 0.030
Identities = 30/77 (38%), Positives = 37/77 (47%), Gaps = 1/77 (1%)
Frame = +2
Query: 187 TSSKGNSP-SPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKT 245
TSS G SP SP P G +S TSP + SP P S T S SS +P T
Sbjct: 815 TSSPGYSPSSPGYSPSSPG-YSPTSPGYSPTSPGYSPTSPGYSPTSPTYSPSSPGYSP-T 988
Query: 246 GRSTTPNSSNRATTSPS 262
+ +P S + + TSPS
Sbjct: 989 SPAYSPTSPSYSPTSPS 1039
Score = 33.9 bits (76), Expect = 0.087
Identities = 29/84 (34%), Positives = 41/84 (48%), Gaps = 7/84 (8%)
Frame = +2
Query: 186 STSSKGNSP-SPSLQPQRVG------AFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSS 238
S +S G SP SP+ P G A+S TSP+ + SP P S + S +S
Sbjct: 917 SPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTS 1096
Query: 239 RSTTPKTGRSTTPNSSNRATTSPS 262
S +P T + +P SS + TSP+
Sbjct: 1097SSYSP-TSPAYSPTSSAYSPTSPA 1165
Score = 32.3 bits (72), Expect = 0.25
Identities = 34/104 (32%), Positives = 42/104 (39%), Gaps = 7/104 (6%)
Frame = +2
Query: 186 STSSKGNSP-SPSLQPQRVG------AFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSS 238
S SS G SP SP P G +S TSP + SP P S S +S
Sbjct: 833 SPSSPGYSPSSPGYSPTSPGYSPTSPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTS 1012
Query: 239 RSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
S +P T S +P S + + TSPS + P A SS
Sbjct: 1013PSYSP-TSPSYSPTSPSYSPTSPSYSPTSSSYSPTSPAYSPTSS 1141
>TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon protein
[imported] - Arabidopsis thaliana, partial (20%)
Length = 1266
Score = 34.7 bits (78), Expect = 0.051
Identities = 28/100 (28%), Positives = 42/100 (42%), Gaps = 3/100 (3%)
Frame = +3
Query: 182 ETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSM---SS 238
+ P + ++ +S P+ RV P+ + K S F S GS SS
Sbjct: 552 QMPLKSETETHSSRPTALKLRVDNIQI-EPSSRSNVVSKHHASMSSFGSSTNGSKRISSS 728
Query: 239 RSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASM 278
R +P T RS+TP+ T PS + PS P A++
Sbjct: 729 RGPSPATSRSSTPSG---RPTLPSTTKSSRPSTPTSRATL 839
>TC86160 weakly similar to PIR|T01605|T01605 phytocyanin At2g44790
[imported] - Arabidopsis thaliana, partial (34%)
Length = 918
Score = 34.3 bits (77), Expect = 0.067
Identities = 35/149 (23%), Positives = 59/149 (39%), Gaps = 8/149 (5%)
Frame = +3
Query: 145 GETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVG 204
G+T ++ KY H DE D +++ + + GNS P + G
Sbjct: 249 GKTFKVGDNLVFKYGSSHQVDEVDESDYKSCTSSNA----IKNYAGGNSKVPLT---KAG 407
Query: 205 AFSFTSPNMA--------KKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPNSSNR 256
F P + K ++ S P S STTP T STTP+++
Sbjct: 408 KIYFICPTLGHCTSTGGMKLEVNVVAASTTPTPSGTPPPTKSPSTTPSTTPSTTPSTT-- 581
Query: 257 ATTSPSNARVRYPSEPRKSASMRLSSDVN 285
++PS PS P+ + ++ +S+ V+
Sbjct: 582 -PSAPSETNSTTPSPPKDNGAVGVSNGVS 665
>AL374845 similar to GP|12247468|gb| PnFL-1 {Ipomoea nil}, partial (25%)
Length = 460
Score = 33.5 bits (75), Expect = 0.11
Identities = 26/93 (27%), Positives = 49/93 (51%)
Frame = +1
Query: 170 HHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFP 229
HH+ + +++ I TP+ + NS S S + A+S SP+++ + SPH P
Sbjct: 49 HHYLSTLQSPIPSTPSPPPTPQNSSSASSNT*PISAYS--SPSLS------HSSSPH--P 198
Query: 230 LSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPS 262
LS ++S S + S+ P+S++ ++ SP+
Sbjct: 199 LSLLSVLTSSSLS-----SSAPSSTSFSSVSPN 282
>TC81882 similar to GP|9759349|dbj|BAB10004.1 contains similarity to unknown
protein~gb|AAF72944.1~gene_id:MAH20.11 {Arabidopsis
thaliana}, partial (5%)
Length = 875
Score = 33.1 bits (74), Expect = 0.15
Identities = 22/77 (28%), Positives = 38/77 (48%), Gaps = 3/77 (3%)
Frame = +2
Query: 182 ETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRST 241
+TPT+ SK ++P P +P ++ +F+ + + R+ PH + S S + T
Sbjct: 170 DTPTTVPSKPSAPKPK-KPPKLLSFADDEIDADNETPRPRSSKPHHHRPKPSSSSSHKIT 346
Query: 242 TPK---TGRSTTPNSSN 255
T K T S +P+ SN
Sbjct: 347 THKNRITSHSPSPSPSN 397
>TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protein
[imported] - Arabidopsis thaliana, partial (70%)
Length = 1259
Score = 32.7 bits (73), Expect = 0.19
Identities = 17/43 (39%), Positives = 27/43 (62%)
Frame = +1
Query: 235 SMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSAS 277
S SSRS++ RS + +SS+R+ + S +R R P PR+ +S
Sbjct: 31 SSSSRSSSRSRSRSRSFSSSSRSPSRSSKSRSRSPPPPRRKSS 159
>AW208110 similar to GP|14715462|d CjMDR1 {Coptis japonica}, partial (11%)
Length = 449
Score = 32.0 bits (71), Expect = 0.33
Identities = 20/41 (48%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Frame = -1
Query: 194 PSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPH-RFPLSRT 233
PSPSLQ ++ F +T P+ DL RT+S H R LSRT
Sbjct: 167 PSPSLQHRKKKLFLYTLPHHLPLDLLPRTISLHLR*ILSRT 45
>TC83980 similar to GP|10177915|dbj|BAB11326. gene_id:MCA23.11~unknown
protein {Arabidopsis thaliana}, partial (36%)
Length = 813
Score = 31.6 bits (70), Expect = 0.43
Identities = 16/60 (26%), Positives = 32/60 (52%)
Frame = +2
Query: 92 DEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVISTPKHHKRYILPVGETLHGT 151
D +V + D R + ++I+TC +DH+ ++Q + K+ Y++ +G + HGT
Sbjct: 509 DGQVLDRINLDRRRNIFGRQIQTCDFVLDHQSVSRQHAAVVPHKNGSVYVIDLG-SAHGT 685
>TC88257 weakly similar to PIR|G96668|G96668 protein F1N19.7 [imported] -
Arabidopsis thaliana, partial (29%)
Length = 1283
Score = 31.2 bits (69), Expect = 0.56
Identities = 17/73 (23%), Positives = 34/73 (46%), Gaps = 2/73 (2%)
Frame = -2
Query: 224 SPHRFPLSRTGSMSSRSTTPKTGRS--TTPNSSNRATTSPSNARVRYPSEPRKSASMRLS 281
+P+ P R+ +++ +T P +S TT N+ + TS R++YP + + +
Sbjct: 1102 APYLTPSIRSNQLTTPTTIPTRNKSNPTTSNTPLSSNTSLHTCRIQYPPKSTRFIKLHQL 923
Query: 282 SDVNNIRDIDQHP 294
I + +HP
Sbjct: 922 IQPTQIPTLHKHP 884
>BQ137346 homologue to GP|15027095|em hypothetical protein LT.09 {Leishmania
major}, partial (0%)
Length = 1101
Score = 30.8 bits (68), Expect = 0.74
Identities = 40/171 (23%), Positives = 65/171 (37%), Gaps = 28/171 (16%)
Frame = +1
Query: 173 RNAVRATIRETPTST------SSKGNSPSPSLQPQRVGAFSFTSPNMA---KKDLEKRTV 223
RN R T ++ T + +G + PS + GA + PN K++
Sbjct: 352 RNKQRTTGKQPKTKAGENARKTKEGQTGEPSGEKPDKGARNRDRPNRGTTGKRNNPPAGG 531
Query: 224 SPHRFPLSRTGSMS--SRSTTPKTGRSTTPNSSNRAT-------------TSPSNARVRY 268
+P R ++T + +R TT R T + +R T TS +
Sbjct: 532 APKRKTRTKTDTTERETRGTTRGRTRRTAEQNRHRHTHDRDTQRRDEREDTSQRKHTQKK 711
Query: 269 PSEPRKSASMRLSSDVNNIRDID----QHPSKSKRLLKSLLSRRKSKKDDT 315
P +P A R + N R D +HP++ K + +R K +DDT
Sbjct: 712 PRQPH--AQKRADRNRRNRRSADADKHKHPTRRKEQQSKMRAREKKTRDDT 858
>TC78685 weakly similar to GP|9757770|dbj|BAB08379.1 gene_id:MOK16.10~unknown
protein {Arabidopsis thaliana}, partial (36%)
Length = 2124
Score = 30.4 bits (67), Expect = 0.96
Identities = 23/68 (33%), Positives = 30/68 (43%)
Frame = -1
Query: 210 SPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYP 269
SP+ L + + H PLS T + ST+P STT S T S + AR
Sbjct: 1791 SPSSFSGHLRNNSSTKHIAPLSNTSDKNLSSTSPAFIFSTT--KSAYCTVSLNQARKSNS 1618
Query: 270 SEPRKSAS 277
S P K +S
Sbjct: 1617 SSPTKGSS 1594
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.128 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,102,088
Number of Sequences: 36976
Number of extensions: 124511
Number of successful extensions: 834
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 811
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 828
length of query: 323
length of database: 9,014,727
effective HSP length: 96
effective length of query: 227
effective length of database: 5,465,031
effective search space: 1240562037
effective search space used: 1240562037
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149127.4


