

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
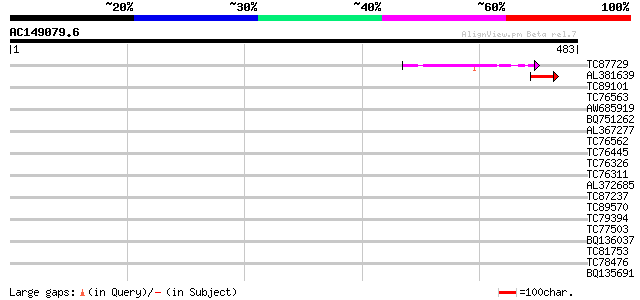
Query= AC149079.6 - phase: 0
(483 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87729 similar to GP|9759461|dbj|BAB10377.1 gene_id:MAF19.15~un... 43 3e-04
AL381639 weakly similar to PIR|T01518|T015 hypothetical protein ... 42 5e-04
TC89101 similar to GP|7268289|emb|CAB78584.1 UFD1 like protein {... 40 0.002
TC76563 homologue to PIR|C29356|C29356 hydroxyproline-rich glyco... 40 0.002
AW685919 homologue to PIR|S25298|S252 extensin (clone Tom J-10) ... 39 0.003
BQ751262 similar to GP|6715100|gb|A versicolorin B synthase {Asp... 39 0.004
AL367277 homologue to PIR|S14974|S149 extensin class I (clone uG... 38 0.008
TC76562 similar to PIR|T11622|T11622 extensin class 1 precursor ... 38 0.008
TC76445 similar to PIR|T07623|T07623 extensin homolog HRGP2 - so... 38 0.008
TC76326 homologue to PIR|C29356|C29356 hydroxyproline-rich glyco... 38 0.008
TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer a... 38 0.008
AL372685 weakly similar to GP|4033606|dbj Extensin {Adiantum cap... 38 0.008
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 38 0.010
TC89570 similar to GP|13936308|gb|AAK40307.1 putative methyl-bin... 38 0.010
TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding... 37 0.013
TC77503 weakly similar to GP|16930417|gb|AAL31894.1 At2g24420/T2... 37 0.013
BQ136037 similar to GP|21322711|em pherophorin-dz1 protein {Volv... 37 0.017
TC81753 similar to PIR|G96752|G96752 unknown protein F28P22.2 [i... 37 0.017
TC78476 homologue to SP|O22437|CHLD_PEA Magnesium-chelatase subu... 37 0.017
BQ135691 similar to PIR|E96636|E966 hypothetical protein T7P1.21... 37 0.022
>TC87729 similar to GP|9759461|dbj|BAB10377.1 gene_id:MAF19.15~unknown protein
{Arabidopsis thaliana}, partial (37%)
Length = 2210
Score = 42.7 bits (99), Expect = 3e-04
Identities = 40/119 (33%), Positives = 58/119 (48%), Gaps = 2/119 (1%)
Frame = +3
Query: 335 PPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERF 394
P ++ KS+FL K+ R+ E +E E ED+EE+E S M++ + +
Sbjct: 1527 PKSIPRKSSFLPPA---KSSRHQVGYPDDERDESEYETEDDEEEERPRSRMRDDDSEAEY 1697
Query: 395 --IEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKK 451
E+ +E E SDEDED G LKQ K++K + + G D SDEG KK
Sbjct: 1698 EDEEEDEQIEQENDASDEDEDEG-LKQKSKNLKRK-----AKKKGFD--SDEGSPPPKK 1850
>AL381639 weakly similar to PIR|T01518|T015 hypothetical protein T10M13.17 -
Arabidopsis thaliana, partial (19%)
Length = 493
Score = 42.0 bits (97), Expect = 5e-04
Identities = 18/24 (75%), Positives = 22/24 (91%)
Frame = +3
Query: 444 EGPDVDKKADEFIAKFREQIRLQR 467
EG +DK+ADEFIAKFR+Q+RLQR
Sbjct: 135 EGEGIDKRADEFIAKFRQQMRLQR 206
>TC89101 similar to GP|7268289|emb|CAB78584.1 UFD1 like protein {Arabidopsis
thaliana}, partial (6%)
Length = 894
Score = 40.4 bits (93), Expect = 0.002
Identities = 30/120 (25%), Positives = 55/120 (45%), Gaps = 3/120 (2%)
Frame = +1
Query: 233 VEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASK 292
+E+++ KE+E A L P R + + QE L + +A++ +
Sbjct: 121 IENQVHNKEQEEAPLLLTKP-RPHTIEVEVHNKDQEEELLLLITPQPDTMEVQAHKEEEQ 297
Query: 293 PSMVKTSSIKFTPSE---SVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRY 349
+++T ++K E S + E++ K+S S PPPPPPPPP++ L+ +
Sbjct: 298 ALLLQTCALKINDKEEEHSPKQGQEEVHMKES---DSLPPPPPPPPPSLHEVEAILEYEF 468
>TC76563 homologue to PIR|C29356|C29356 hydroxyproline-rich glycoprotein
(clone Hyp2.13) - kidney bean (fragment), partial (34%)
Length = 507
Score = 40.4 bits (93), Expect = 0.002
Identities = 16/26 (61%), Positives = 18/26 (68%)
Frame = +2
Query: 317 IKKKSFYYKSYPPPPPPPPPTMFYKS 342
I K +YYKS PPP P PPP +YKS
Sbjct: 224 IPKTPYYYKSPPPPSPSPPPPYYYKS 301
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 383 YYYKSPPPPSPSPPPPYYYKS 445
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 287 YYYKSPPPPSPSPPPPYYYKS 349
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 335 YYYKSPPPPSPSPPPPYYYKS 397
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 143 YYYKSPPPPSPSPPPPYYYKS 205
Score = 31.2 bits (69), Expect = 0.93
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P P +YKS
Sbjct: 191 YYYKSPPPPSPIPKTPYYYKS 253
Score = 30.8 bits (68), Expect = 1.2
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P P +YKS
Sbjct: 431 YYYKSPPPPSPIPHTPYYYKS 493
>AW685919 homologue to PIR|S25298|S252 extensin (clone Tom J-10) - tomato,
partial (23%)
Length = 663
Score = 39.3 bits (90), Expect = 0.003
Identities = 15/24 (62%), Positives = 17/24 (70%)
Frame = +1
Query: 319 KKSFYYKSYPPPPPPPPPTMFYKS 342
K +YYKS PPP P PPP +YKS
Sbjct: 1 KTPYYYKSPPPPSPSPPPPYYYKS 72
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 106 YYYKSPPPPSPSPPPPYYYKS 168
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 154 YYYKSPPPPSPSPPPPYYYKS 216
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 58 YYYKSPPPPSPSPPPPYYYKS 120
Score = 31.2 bits (69), Expect = 0.93
Identities = 13/25 (52%), Positives = 15/25 (60%), Gaps = 6/25 (24%)
Frame = +1
Query: 322 FYYKSYPPPPP------PPPPTMFY 340
+YYKS PPP P PPPP +Y
Sbjct: 202 YYYKSPPPPAPVYKYNSPPPPVHYY 276
>BQ751262 similar to GP|6715100|gb|A versicolorin B synthase {Aspergillus
parasiticus}, partial (11%)
Length = 710
Score = 38.9 bits (89), Expect = 0.004
Identities = 34/115 (29%), Positives = 51/115 (43%), Gaps = 8/115 (6%)
Frame = +3
Query: 231 GLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENA 290
GL+ ++ +A + SP+ +RSAS R P+ E + S P + ++A
Sbjct: 318 GLLRSRMPTMPSRSAHISSPLS--TRSASPR---PRTSTAESSWARSTAPTPSPQRRKSA 482
Query: 291 S--------KPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPT 337
S KPS +TS + TPS S ++S K+ S PP P PPT
Sbjct: 483 SRARRRSSPKPSAARTS--RSTPSPSPRRSSSTATKRPP---ASSSPPTPASPPT 632
>AL367277 homologue to PIR|S14974|S149 extensin class I (clone uG-18) -
tomato (fragment), partial (46%)
Length = 254
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 1 YYYKSPPPPSPSPPPPYYYKS 63
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 49 YYYKSPPPPSPSPPPPYYYKS 111
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 97 YYYKSPPPPSPSPPPPYYYKS 159
Score = 33.5 bits (75), Expect = 0.19
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = +1
Query: 317 IKKKSFYYKSYPPPPPPPPPTMFYK 341
I + YKS PPP P PPP +YK
Sbjct: 178 ISHPPYNYKSPPPPSPSPPPPYYYK 252
>TC76562 similar to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 1391
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 14 YYYKSPPPPSPSPPPPYYYKS 76
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 110 YYYKSPPPPSPSPPPPYYYKS 172
Score = 32.3 bits (72), Expect = 0.42
Identities = 32/117 (27%), Positives = 46/117 (38%), Gaps = 9/117 (7%)
Frame = +2
Query: 237 LKQKEEENAVLPS-PIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSM 295
LK K++ VL + P + S+ +PK K+ P + N S P
Sbjct: 614 LKSKDKYEVVLKAKPFAYASKKHFKECEKPKPSPTPYYYKSPPPPSPVYKYN---SPPPP 784
Query: 296 VKTSSIKFTPSESVAKNSEDLIKKKS--FYYKSYPPPPP------PPPPTMFYKSTF 344
V S +T K+ +K +YYKS PPP P PPPP +Y +
Sbjct: 785 VHYYSPPYT-----YKSPPPPVKAAPTPYYYKSPPPPAPVYKYNSPPPPVHYYSPPY 940
Score = 31.6 bits (70), Expect = 0.71
Identities = 13/29 (44%), Positives = 16/29 (54%), Gaps = 6/29 (20%)
Frame = +2
Query: 322 FYYKSYPPPPP------PPPPTMFYKSTF 344
+YYKS PPP P PPPP +Y +
Sbjct: 986 YYYKSPPPPAPVYKYNSPPPPVHYYSPPY 1072
Score = 30.8 bits (68), Expect = 1.2
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P P +YKS
Sbjct: 62 YYYKSPPPPSPIPHTPYYYKS 124
Score = 30.8 bits (68), Expect = 1.2
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P P +YKS
Sbjct: 158 YYYKSPPPPSPIPHTPYYYKS 220
Score = 28.5 bits (62), Expect = 6.0
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPPP PP +Y S
Sbjct: 206 YYYKS-PPPPKVLPPPYYYNS 265
>TC76445 similar to PIR|T07623|T07623 extensin homolog HRGP2 - soybean
(fragment), partial (76%)
Length = 821
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 68 YYYKSPPPPSPSPPPPYYYKS 130
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 116 YYYKSPPPPSPSPPPPYYYKS 178
Score = 35.0 bits (79), Expect = 0.064
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP Y+S
Sbjct: 164 YYYKSPPPPSPSPPPPYHYQS 226
Score = 35.0 bits (79), Expect = 0.064
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PP +YKS
Sbjct: 20 YYYKSPPPPSPSPPSPYYYKS 82
Score = 32.7 bits (73), Expect = 0.32
Identities = 13/20 (65%), Positives = 13/20 (65%)
Frame = +2
Query: 323 YYKSYPPPPPPPPPTMFYKS 342
YYKS PPP P PPP Y S
Sbjct: 263 YYKSPPPPSPSPPPPYHYVS 322
>TC76326 homologue to PIR|C29356|C29356 hydroxyproline-rich glycoprotein
(clone Hyp2.13) - kidney bean (fragment), partial (32%)
Length = 777
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 43 YYYKSPPPPSPSPPPPYYYKS 105
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 91 YYYKSPPPPSPSPPPPYYYKS 153
Score = 35.0 bits (79), Expect = 0.064
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP Y+S
Sbjct: 139 YYYKSPPPPSPSPPPPYHYQS 201
Score = 34.3 bits (77), Expect = 0.11
Identities = 14/26 (53%), Positives = 15/26 (56%)
Frame = +1
Query: 317 IKKKSFYYKSYPPPPPPPPPTMFYKS 342
I +YYKS PPP P PPP Y S
Sbjct: 220 ISHPPYYYKSPPPPSPSPPPPYHYVS 297
Score = 29.3 bits (64), Expect = 3.5
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = +1
Query: 328 PPPPPPPPPTMFYKS 342
PPP P PPP +YKS
Sbjct: 13 PPPSPSPPPPYYYKS 57
>TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer arietinum},
partial (77%)
Length = 973
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 77 YYYKSPPPPSPSPPPPYYYKS 139
Score = 36.6 bits (83), Expect = 0.022
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +Y+S
Sbjct: 125 YYYKSPPPPSPSPPPPYYYQS 187
Score = 35.8 bits (81), Expect = 0.038
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YY S PPP P PPP +YKS
Sbjct: 29 YYYHSPPPPSPSPPPPYYYKS 91
Score = 32.0 bits (71), Expect = 0.54
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
++Y S PPP P P PT YKS
Sbjct: 314 YHYTSPPPPSPAPAPTYIYKS 376
Score = 28.9 bits (63), Expect = 4.6
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
++YKS PPP PPP Y S
Sbjct: 221 YHYKSPPPPTASPPPPYHYVS 283
Score = 28.5 bits (62), Expect = 6.0
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YY+S PPP P P YKS
Sbjct: 173 YYYQSPPPPSPTPHTPYHYKS 235
>AL372685 weakly similar to GP|4033606|dbj Extensin {Adiantum
capillus-veneris}, partial (49%)
Length = 407
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 299 YYYKSPPPPSPSPPPPYYYKS 361
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 251 YYYKSPPPPSPSPPPPYYYKS 313
Score = 36.6 bits (83), Expect = 0.022
Identities = 13/20 (65%), Positives = 15/20 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYK 341
+YYKS PPP P PPP +YK
Sbjct: 347 YYYKSPPPPSPSPPPPYYYK 406
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 37.7 bits (86), Expect = 0.010
Identities = 61/325 (18%), Positives = 113/325 (34%), Gaps = 1/325 (0%)
Frame = +1
Query: 121 DEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKP 180
+E H ES++ + + ++ +K + + L+N +DE+ S ENE+
Sbjct: 1 EEKSHLENEESKETGESSEEKSHLENEESKETGESSEEKSHLENEENKDEEKSKQENEE- 177
Query: 181 LLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEG-LVEDKLKQ 239
+ ++ ++ +SQ + +K K N + + G + E+K KQ
Sbjct: 178 -IKDGEKIQQENEENKDEEKSQQENEENKDEEKSQQENELKKNEGGEKETGEITEEKSKQ 354
Query: 240 KEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTS 299
+ EE + + S E Q + K++ EN + S T
Sbjct: 355 ENEETS--------ETNSKDKENEESNQNG----------SDAKEQVGENHEQDSKQGTE 480
Query: 300 SIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSM 359
T E K D IK+ + S+ + + GEKN
Sbjct: 481 ETNGT--EGGEKEEHDKIKEDT--------------------SSDNQVQDGEKNNEAREE 594
Query: 360 GKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQ 419
S + ++ D +E T ++ +K F E + ET D + + Q
Sbjct: 595 NYSGDNASSAVVDNKSQESSNKTEEQFDKKEKNEF-ELESQKNSNETTESTDSTITQNSQ 771
Query: 420 SVKSVKTEEPCSSSLSGGIDTVSDE 444
+S K + + G + SDE
Sbjct: 772 GNESEKDQAQTENDTPKGSASESDE 846
>TC89570 similar to GP|13936308|gb|AAK40307.1 putative methyl-binding domain
protein MBD105 {Zea mays}, partial (9%)
Length = 728
Score = 37.7 bits (86), Expect = 0.010
Identities = 43/216 (19%), Positives = 80/216 (36%), Gaps = 8/216 (3%)
Frame = +3
Query: 247 LPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPS 306
LP+P W + + PK+ I + K++ + S +
Sbjct: 150 LPAPSGWTKKFFPKKSGTPKKNEIVFTSPTGEEIRTKRQLEKYLKTNGGPNISEFDWGTG 329
Query: 307 ESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEG 366
E+ ++S + K K+ + P P PP + + + ++ + ++E
Sbjct: 330 ETPRRSSRIVEKVKA----APPAEPKTDPPKKRSRKSSASKKEASEDEAEETKDVEMQEA 497
Query: 367 TNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKT 426
K+D+D E +EK+V+ E E+ ED DV K+S++ +T
Sbjct: 498 GETKDDKDVE------------------LEKKVVNENEDKKGAEDADV---KESIQPGET 614
Query: 427 EEPCSSSLSGG--------IDTVSDEGPDVDKKADE 454
+ S+ + G ID EG V + DE
Sbjct: 615 TDEGKSNTADGDLQASKENIDDKGAEGSGVVQNKDE 722
>TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding protein -
Arabidopsis thaliana, partial (74%)
Length = 1909
Score = 37.4 bits (85), Expect = 0.013
Identities = 16/56 (28%), Positives = 32/56 (56%)
Frame = +2
Query: 365 EGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQS 420
+G N++E+ EEE EY ++E+ ++E E+ +E E DE+++ + K +
Sbjct: 245 DGDNDQEESSEEEVEYEEVEVEEEVEEEEEEEEEEEVEEESKPLDEEDEADKKKHA 412
>TC77503 weakly similar to GP|16930417|gb|AAL31894.1 At2g24420/T28I24.15
{Arabidopsis thaliana}, partial (54%)
Length = 1787
Score = 37.4 bits (85), Expect = 0.013
Identities = 37/147 (25%), Positives = 71/147 (48%), Gaps = 10/147 (6%)
Frame = +2
Query: 342 STFLKSRYGEKNGRNMSMGKSIEEGTNEKEDED----EEEKEYST--SSMQEQTDKERFI 395
S+ LK + + N + S+ I E T E + +D E+EK + SS+Q ++ +
Sbjct: 206 SSALKIQLDQLNSKIQSLESQISEKTQELKKKDQVIAEKEKLFQDKLSSIQSLQNEVASL 385
Query: 396 EKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSS----LSGGIDTVSDEGPDVDKK 451
+K+ L+AEE GEL++ V +K+E +S + + + D++ K
Sbjct: 386 QKKGSLDAEEQVGKAYARAGELQKQVDKLKSELEAQNSEKVNWESRVAKLEKKIHDLNSK 565
Query: 452 ADEFIAKFREQIRLQRIESIKRSTRVA 478
++ + K E+ + Q I +R+ +VA
Sbjct: 566 LED-VQKINEEQKKQ-IRKTERALKVA 640
>BQ136037 similar to GP|21322711|em pherophorin-dz1 protein {Volvox carteri
f. nagariensis}, partial (5%)
Length = 712
Score = 37.0 bits (84), Expect = 0.017
Identities = 14/23 (60%), Positives = 17/23 (73%)
Frame = +1
Query: 318 KKKSFYYKSYPPPPPPPPPTMFY 340
KK F++ S PPPPPPPPP F+
Sbjct: 301 KKCFFFFFSPPPPPPPPPPHRFF 369
Score = 31.6 bits (70), Expect = 0.71
Identities = 10/19 (52%), Positives = 13/19 (67%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFY 340
F++ PPPPPPPP F+
Sbjct: 316 FFFSPPPPPPPPPPHRFFF 372
Score = 31.2 bits (69), Expect = 0.93
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
F+ + PPPPPPPP F+ S
Sbjct: 317 FFSRHLPPPPPPPPIDFFFFS 379
>TC81753 similar to PIR|G96752|G96752 unknown protein F28P22.2 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 1273
Score = 37.0 bits (84), Expect = 0.017
Identities = 50/234 (21%), Positives = 89/234 (37%), Gaps = 38/234 (16%)
Frame = +3
Query: 158 QVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSS 217
+V+ ++ SV D S ++ P L P+ +L+ + + +S S++ ++ T+ FSS
Sbjct: 21 EVETVEKSVIND--SVVENSQPPPLQPIPALRKKKT------RSASMENVAVVTSAEFSS 176
Query: 218 NSFNRVRNYAEFEGLVEDKLKQKEEEN------AVLPSPIPWRSRSASARMMEPK----- 266
N + K+ + E N A SP+P + + +M K
Sbjct: 177 LVPEPELNPHRQPAARKKKITSESEVNTAAVTSAEFSSPVPEQELNPQLPVMMTKGVRGN 356
Query: 267 ------QEAIEKALKASMVME-------------------PKQEANENASKPSMVKTSSI 301
+AIEK +E P ++ NA+K +
Sbjct: 357 FKRTNQPKAIEKLEDNDFFVENYLPQTVPQRSMAERKTGIPLKKKRGNATKEFLASLRGK 536
Query: 302 KFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPP-PTM-FYKSTFLKSRYGEKN 353
K +N E + + S PPPPPPPP P++ F + + + +KN
Sbjct: 537 KKKQRSKSVENFETIQNSQPSPLVSQPPPPPPPPSPSVGFSQPVHFEQKQTQKN 698
>TC78476 homologue to SP|O22437|CHLD_PEA Magnesium-chelatase subunit chlD
chloroplast precursor (Mg- protoporphyrin IX chelatase),
partial (68%)
Length = 1596
Score = 37.0 bits (84), Expect = 0.017
Identities = 27/108 (25%), Positives = 41/108 (37%)
Frame = +2
Query: 285 EANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTF 344
E E + K + P V N + ++ PPPPPPPP
Sbjct: 392 EGREKVYADDLKKAVELVILPRSIVTDNPPE--------QQNQPPPPPPPPQN------- 526
Query: 345 LKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKE 392
+ E+N EE E+E+E+E++K+ QEQ +E
Sbjct: 527 -QESNEEQN----------EEEDKEEEEEEEDDKDEEKEEQQEQLPEE 637
>BQ135691 similar to PIR|E96636|E966 hypothetical protein T7P1.21 [imported]
- Arabidopsis thaliana, partial (4%)
Length = 1215
Score = 36.6 bits (83), Expect = 0.022
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = +1
Query: 328 PPPPPPPPPTMFYKSTFLKSRYG 350
PPPPPPPPP+ F+ F R+G
Sbjct: 10 PPPPPPPPPSPFFALFFFGPRFG 78
Score = 30.8 bits (68), Expect = 1.2
Identities = 12/22 (54%), Positives = 14/22 (63%)
Frame = +3
Query: 328 PPPPPPPPPTMFYKSTFLKSRY 349
PPPPPPPPP + S L S +
Sbjct: 9 PPPPPPPPPLPLFCSFLLWSPF 74
Score = 30.0 bits (66), Expect = 2.1
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = +3
Query: 328 PPPPPPPPPTMFYKSTFL 345
PPPPPPPPP + +FL
Sbjct: 6 PPPPPPPPPPLPLFCSFL 59
Score = 28.9 bits (63), Expect = 4.6
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +3
Query: 328 PPPPPPPPP 336
PPPPPPPPP
Sbjct: 3 PPPPPPPPP 29
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,488,765
Number of Sequences: 36976
Number of extensions: 221974
Number of successful extensions: 8910
Number of sequences better than 10.0: 273
Number of HSP's better than 10.0 without gapping: 2927
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5987
length of query: 483
length of database: 9,014,727
effective HSP length: 100
effective length of query: 383
effective length of database: 5,317,127
effective search space: 2036459641
effective search space used: 2036459641
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149079.6


