

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.14 - phase: 0
(309 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
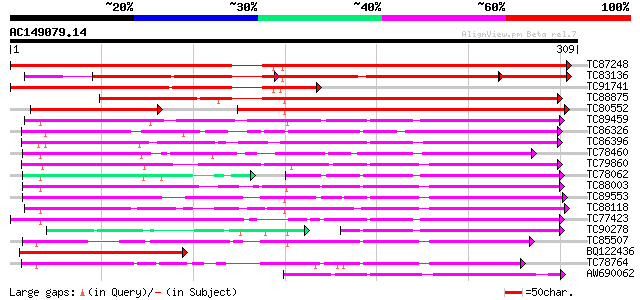
Score E
Sequences producing significant alignments: (bits) Value
TC87248 similar to GP|7594580|emb|CAB88075.1 hypothetical protei... 548 e-157
TC83136 similar to GP|7594580|emb|CAB88075.1 hypothetical protei... 342 e-110
TC91741 similar to GP|20303569|gb|AAM18997.1 putative gibberelli... 239 7e-64
TC88875 similar to PIR|G86472|G86472 probable hyoscyamine 6-diox... 228 2e-60
TC80552 similar to PIR|G86472|G86472 probable hyoscyamine 6-diox... 176 8e-45
TC89459 weakly similar to GP|9280317|dbj|BAB01696.1 oxylase-like... 115 2e-26
TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoantho... 105 2e-23
TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin ... 104 4e-23
TC78460 similar to GP|21554579|gb|AAM63621.1 flavonol synthase-l... 104 4e-23
TC79860 homologue to PIR|T06533|T06533 probable gibberellin 20-o... 98 5e-21
TC78062 weakly similar to GP|9280317|dbj|BAB01696.1 oxylase-like... 81 5e-21
TC88003 weakly similar to PIR|T04184|T04184 hypothetical protein... 96 2e-20
TC89553 weakly similar to SP|P24397|HY6H_HYONI Hyoscyamine 6-dio... 96 2e-20
TC88118 weakly similar to PIR|T09683|T09683 gibberellin 7-oxidas... 94 7e-20
TC77423 weakly similar to PIR|D96569|D96569 probable oxidoreduct... 91 4e-19
TC90278 weakly similar to GP|9280318|dbj|BAB01697.1 oxidase-like... 67 5e-18
TC85507 homologue to SP|P31239|ACCO_PEA 1-aminocyclopropane-1-ca... 87 8e-18
BQ122436 weakly similar to PIR|G86472|G86 probable hyoscyamine 6... 87 8e-18
TC78764 similar to PIR|C96802|C96802 hypothetical protein F2P24.... 87 1e-17
AW690062 weakly similar to GP|7268917|emb gibberellin 20-oxidase... 83 2e-16
>TC87248 similar to GP|7594580|emb|CAB88075.1 hypothetical protein
{Arabidopsis thaliana}, partial (82%)
Length = 1271
Score = 548 bits (1413), Expect = e-157
Identities = 276/312 (88%), Positives = 280/312 (89%), Gaps = 6/312 (1%)
Frame = +1
Query: 1 MAGTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFS 60
MAGTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFS
Sbjct: 52 MAGTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFS 231
Query: 61 LPIEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGV 120
LPIEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSN +
Sbjct: 232 LPIEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNEL 411
Query: 121 FSFFPAMTLQNYSQIGDPQWNP----YFGKY--SGKELLSLIALSLNLDENYFEKISALN 174
P W P F K +GKELLSLIALSLNLDENYFEKISALN
Sbjct: 412 L----------------PNWRPTMESLFWKILSAGKELLSLIALSLNLDENYFEKISALN 543
Query: 175 KPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDV 234
KPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDV
Sbjct: 544 KPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDV 723
Query: 235 SHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCS 294
SHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCS
Sbjct: 724 SHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCS 903
Query: 295 ESSPPRYRKVLS 306
ESSPPR+ + S
Sbjct: 904 ESSPPRFPPIRS 939
>TC83136 similar to GP|7594580|emb|CAB88075.1 hypothetical protein
{Arabidopsis thaliana}, partial (59%)
Length = 1090
Score = 342 bits (878), Expect(2) = e-110
Identities = 177/230 (76%), Positives = 189/230 (81%), Gaps = 6/230 (2%)
Frame = +1
Query: 46 DFIKQAFDQSANFFSLPIEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLT 105
DF+KQAF+QSA FFSLPIEDKIKLNRKEYRGYTPLYAEKLDPS LSKGDPKESYYIG L
Sbjct: 61 DFMKQAFEQSAKFFSLPIEDKIKLNRKEYRGYTPLYAEKLDPS-LSKGDPKESYYIGSLE 237
Query: 106 DTTSVRLNQWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKY------SGKELLSLIALS 159
DTTSVRLNQWPS+ + P W P +GKELLSLIALS
Sbjct: 238 DTTSVRLNQWPSDKLL----------------PNWKPTMESLFWKTLSAGKELLSLIALS 369
Query: 160 LNLDENYFEKISALNKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQ 219
LNLDE+YF+KISA NKP+AFLRLLRYPGELG CGASAHSDYGMITLL+TNGVPGLQ
Sbjct: 370 LNLDEDYFQKISASNKPDAFLRLLRYPGELG----CCGASAHSDYGMITLLITNGVPGLQ 537
Query: 220 ICKEKLKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGK 269
ICKEKLKQPQVWEDVSHVEGAIIVNIGD+MERWTNC+Y+STLHRVMPTGK
Sbjct: 538 ICKEKLKQPQVWEDVSHVEGAIIVNIGDIMERWTNCVYQSTLHRVMPTGK 687
Score = 75.5 bits (184), Expect(2) = e-110
Identities = 31/39 (79%), Positives = 36/39 (91%)
Frame = +2
Query: 268 GKERYSVAFFMDPPSDCVVECFESCCSESSPPRYRKVLS 306
GKERYSVAF+MDPPSDC+V+CFESCCSESSPPR+ + S
Sbjct: 683 GKERYSVAFYMDPPSDCIVKCFESCCSESSPPRFPPIRS 799
Score = 44.7 bits (104), Expect = 5e-05
Identities = 42/139 (30%), Positives = 59/139 (42%)
Frame = +2
Query: 9 LPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKIK 68
LPII LSSPDR+S+A SI Q + + +FF ++ K+
Sbjct: 2 LPIIYLSSPDRLSSANSIRQI-----------------L*SKRSSRVQSFFHFQLKTKLN 130
Query: 69 LNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFFPAMT 128
R + L +L+ PK + ++ L W VF +
Sbjct: 131 *TA---RSTEVILLSML--RNLTLHSPK-------VIQKRAITLALWKIPLVFV*INGL- 271
Query: 129 LQNYSQIGDPQWNPYFGKY 147
L NY QIG+PQWNPYFGK+
Sbjct: 272 LINYYQIGNPQWNPYFGKH 328
>TC91741 similar to GP|20303569|gb|AAM18997.1 putative gibberellin
20-oxidase {Oryza sativa (japonica cultivar-group)},
partial (14%)
Length = 501
Score = 239 bits (611), Expect = 7e-64
Identities = 128/176 (72%), Positives = 141/176 (79%), Gaps = 6/176 (3%)
Frame = +2
Query: 1 MAGTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFS 60
MAGTAT++LPII+LSSPDR+S+A SI QACVEYGFFYLVNHGVD+DF+KQAF+QSA FFS
Sbjct: 23 MAGTATVNLPIINLSSPDRLSSANSIRQACVEYGFFYLVNHGVDKDFMKQAFEQSAKFFS 202
Query: 61 LPIEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGV 120
LPIEDKIKLNRKEYRGYTPLYAEKLDP SLSKGDPKESYYIG L DTTSVRLNQWPS+ +
Sbjct: 203 LPIEDKIKLNRKEYRGYTPLYAEKLDP-SLSKGDPKESYYIGSLEDTTSVRLNQWPSDKL 379
Query: 121 FSFFPAMTLQNYSQIGDPQWNP----YFGK--YSGKELLSLIALSLNLDENYFEKI 170
P W P F K +GKELLSLIALSLNLDE+YF+KI
Sbjct: 380 L----------------PNWKPTMESLFWKTLSAGKELLSLIALSLNLDEDYFQKI 499
>TC88875 similar to PIR|G86472|G86472 probable hyoscyamine 6-dioxygenase
hydroxylase [imported] - Arabidopsis thaliana, partial
(82%)
Length = 1085
Score = 228 bits (582), Expect = 2e-60
Identities = 119/266 (44%), Positives = 169/266 (62%), Gaps = 14/266 (5%)
Frame = +2
Query: 50 QAFDQSANFFSLPIEDKIKLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTT 108
+ F QS FF+LP+ +K+KL R + ++GYTP+ E LDP + GD KE YYIG + D
Sbjct: 92 KVFAQSNTFFTLPLNEKMKLLRNDSHKGYTPVLDEILDPQNQLHGDYKEGYYIG-VEDND 268
Query: 109 SVRL------NQWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYS------GKELLSLI 156
S+ NQWP++ V P W +Y GK L +I
Sbjct: 269 SLHHKPFYGPNQWPASDVL----------------PGWRETMEEYQRRALEVGKALARII 400
Query: 157 ALSLNLDENYFEKISALNKPEAFLRLLRYPGELG-PNEEICGASAHSDYGMITLLLTNGV 215
AL+L LD N+F+K L +P A LRLL Y ++ P++ + A AH+DYG+ITLL T+ V
Sbjct: 401 ALALGLDANFFDKPETLREPIAILRLLHYQDKVSDPSQGLYAAGAHTDYGLITLLATDDV 580
Query: 216 PGLQICKEKLKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVA 275
GLQICK K +PQ+WEDV+ ++GA IVN+GDM+ERW+NC ++STLHRV+ G+ERYS+A
Sbjct: 581 QGLQICKNKDAKPQIWEDVTPLKGAFIVNLGDMLERWSNCTFKSTLHRVIGNGQERYSIA 760
Query: 276 FFMDPPSDCVVECFESCCSESSPPRY 301
+F++P DC+VEC +C S+ +PP++
Sbjct: 761 YFLEPGHDCLVECLPTCKSDENPPKF 838
Score = 39.3 bits (90), Expect = 0.002
Identities = 13/34 (38%), Positives = 23/34 (67%)
Frame = +1
Query: 26 IHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFF 59
+ Q+C + G+FYL+NHG+ +DF+ + F + F
Sbjct: 19 LKQSCTDSGWFYLINHGISQDFMDEGFCPEQHIF 120
>TC80552 similar to PIR|G86472|G86472 probable hyoscyamine 6-dioxygenase
hydroxylase [imported] - Arabidopsis thaliana, partial
(92%)
Length = 1307
Score = 176 bits (447), Expect = 8e-45
Identities = 89/189 (47%), Positives = 131/189 (69%), Gaps = 8/189 (4%)
Frame = +3
Query: 125 PAMTLQN-YSQIGDPQWNPYFGKYS------GKELLSLIALSLNLDENYFEKISALNKPE 177
P M L N QI P W K+ GK + +IAL+L+LD ++F+K L +
Sbjct: 333 PFMGLINGLHQILLPGWRETMEKFHREAL*VGKAVGKIIALALDLDADFFDKPEMLGESI 512
Query: 178 AFLRLLRYPGELG-PNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSH 236
A LRLL Y G++ P + + GA AH+DYG+ITLL T+ V GLQICK++ +PQ WEDV+
Sbjct: 513 ATLRLLHYGGQVSDPLKGLFGAGAHTDYGLITLLATDHVSGLQICKDRDAKPQKWEDVAP 692
Query: 237 VEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSES 296
++GA +VN+GDM+ERW+N +++STLHRV+ G+ERYS+A+F++P DC+VEC +C S++
Sbjct: 693 LKGAFVVNLGDMLERWSNGVFKSTLHRVLGNGQERYSIAYFLEPSHDCLVECLPTCKSDT 872
Query: 297 SPPRYRKVL 305
+PP+Y +L
Sbjct: 873 NPPKYPPIL 899
Score = 78.2 bits (191), Expect = 4e-15
Identities = 34/73 (46%), Positives = 54/73 (73%), Gaps = 1/73 (1%)
Frame = +1
Query: 12 IDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKIKLNR 71
IDL +P+ + + QAC++ GFFY+VNHG+ E+F+ + F+QS FF+LP+++K+K+ R
Sbjct: 16 IDLPNPNINQSVNLLKQACLDSGFFYVVNHGISEEFMDEVFEQSKRFFTLPLKEKMKILR 195
Query: 72 KE-YRGYTPLYAE 83
E +RGYTP+ E
Sbjct: 196NEKHRGYTPVLDE 234
>TC89459 weakly similar to GP|9280317|dbj|BAB01696.1 oxylase-like protein
{Arabidopsis thaliana}, partial (70%)
Length = 1302
Score = 115 bits (289), Expect = 2e-26
Identities = 100/312 (32%), Positives = 142/312 (45%), Gaps = 18/312 (5%)
Frame = +3
Query: 9 LPIIDLS--------SPDRIST-AKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFF 59
+P IDLS +P I K I A E+GFF + NHGV ++ + S FF
Sbjct: 138 IPEIDLSPILHHAVPNPSDIENLVKEIGSASKEWGFFQVTNHGVPLSLRQRLEEASRLFF 317
Query: 60 SLPIEDKIKLNRKEYR--GYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPS 117
+ +E+K K+ R E GY K + D KE + T R +
Sbjct: 318 AQSLEEKKKVARDEVNPTGYYDTEHTK------NVRDWKEVFDFLSQDPTLVPRSSDEHD 479
Query: 118 NGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGK------ELLSLIALSLNLDENYFEKIS 171
+GV + N S PQ+ +Y + LL LIALSL L+ FE+
Sbjct: 480 DGVIQW------TNQSPQYPPQFRAILEEYIQEVEKLAYRLLELIALSLGLEAKRFEEF- 638
Query: 172 ALNKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVW 231
+F+R YP P+ + G H D G +T+L + V GL++ K K Q W
Sbjct: 639 -FKYQTSFIRFNHYPPCPYPHLAL-GVGRHKDAGALTILAQDEVGGLEV---KRKSDQQW 803
Query: 232 EDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTG-KERYSVAFFMDPPSDCVVECFE 290
V A I+N+GD+++ W+N Y S HRVM KER+S+ FF P D VV+ E
Sbjct: 804 VLVKPTPDAYIINVGDIIQVWSNDAYESVEHRVMVNSEKERFSIPFFFFPAHDTVVKPLE 983
Query: 291 SCCSESSPPRYR 302
+E +PP+YR
Sbjct: 984 ELTNEENPPKYR 1019
>TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoanthocyanidin
dioxygenase {Arabidopsis thaliana}, partial (81%)
Length = 1686
Score = 105 bits (262), Expect = 2e-23
Identities = 91/306 (29%), Positives = 148/306 (47%), Gaps = 11/306 (3%)
Frame = +2
Query: 7 LSLPIIDLSSPD------RISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFS 60
+++PIIDL + S K I AC ++GFF +VNHGV D + +A + FF
Sbjct: 392 INIPIIDLGGLNGDDLDVHASILKQISDACRDWGFFQIVNHGVSPDLMDKARETWRQFFH 571
Query: 61 LPIEDKIKLNRKEYRGYTPLYAEKLDPSSLSKG---DPKESYYIGPLTDTTSVR-LNQWP 116
LP+E K ++Y Y + KG D + Y++ L SV+ N+WP
Sbjct: 572 LPMEAK-----QQYANSPTTYEGYGSRLGVEKGAILDWSDYYFLHYL--PVSVKDCNKWP 730
Query: 117 SNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKP 176
++ Q+ ++ D ++ K SG+ L+ ++L+L L+E + +
Sbjct: 731 ASP----------QSCREVFD-EYGKELVKLSGR-LMKALSLNLGLEEKILQNAFGGEEI 874
Query: 177 EAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSH 236
A +R+ YP P E G S+HSD G +T+LL P Q+ ++++ W V+
Sbjct: 875 GACMRVNFYPKCPRP-ELTLGLSSHSDPGGMTMLL----PDDQVAGLQVRKFDNWITVNP 1039
Query: 237 VEGAIIVNIGDMMERWTNCLYRSTLHRV-MPTGKERYSVAFFMDPPSDCVVECFESCCSE 295
IVNIGD ++ +N Y+S HRV + + +ER S+AFF +P SD +E + +
Sbjct: 1040ARHGFIVNIGDQIQVLSNATYKSVEHRVIVNSDQERLSLAFFYNPRSDIPIEPLKQLITP 1219
Query: 296 SSPPRY 301
P Y
Sbjct: 1220ERPALY 1237
>TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin
dioxygenase-like protein {Arabidopsis thaliana}, partial
(82%)
Length = 1326
Score = 104 bits (260), Expect = 4e-23
Identities = 88/306 (28%), Positives = 152/306 (48%), Gaps = 11/306 (3%)
Frame = +2
Query: 7 LSLPIIDL---SSPD---RISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFS 60
+++P+IDL SS D R + K + +AC E+GFF +VNHG+ + ++ A + FF+
Sbjct: 248 INIPVIDLEHLSSEDPVLRETVLKRVSEACREWGFFQVVNHGISHELMESAKEVWREFFN 427
Query: 61 LPIEDKIKL--NRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSN 118
LP+E K + + Y G Y +L + D + +++ + + + +WP+
Sbjct: 428 LPLEVKEEFANSPSTYEG----YGSRLGVKKGAILDWSDYFFLHSMPPSLRNQA-KWPAT 592
Query: 119 GVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENY-FEKISALNKPE 177
P+ + ++ G+ G +L L++ +L L E+Y N+
Sbjct: 593 ------PSSLRKIIAEYGEEVVK------LGGRMLELMSTNLGLKEDYLMNAFGGENELG 736
Query: 178 AFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNG-VPGLQICKEKLKQPQVWEDVSH 236
A LR+ YP P+ + G S HSD G +T+LL + V GLQ+ ++ W V
Sbjct: 737 ACLRVNFYPKCPQPDLTL-GLSPHSDPGGMTILLPDDFVSGLQV-----RKGNDWITVKP 898
Query: 237 VEGAIIVNIGDMMERWTNCLYRSTLHRVMPTG-KERYSVAFFMDPPSDCVVECFESCCSE 295
V A I+NIGD ++ +N +Y+S HRV+ K+R S+A F +P SD ++E + ++
Sbjct: 899 VPNAFIINIGDQIQVLSNAIYKSVEHRVIVNSIKDRVSLAMFYNPKSDLLIEPAKELVTK 1078
Query: 296 SSPPRY 301
P Y
Sbjct: 1079ERPALY 1096
>TC78460 similar to GP|21554579|gb|AAM63621.1 flavonol synthase-like protein
{Arabidopsis thaliana}, partial (86%)
Length = 1442
Score = 104 bits (260), Expect = 4e-23
Identities = 92/309 (29%), Positives = 138/309 (43%), Gaps = 29/309 (9%)
Frame = +2
Query: 8 SLPIIDLS-------------SPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQ 54
S+PIID+S P + + +AC E GFFY+ HG+ + +K D
Sbjct: 158 SIPIIDISPLLAKSDDPKMAEDPGVLDVVAQLDKACTEAGFFYVKGHGIPDSLLKGVRDI 337
Query: 55 SANFFSLPIEDKIKLN---RKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTS-- 109
+ FF LP E+K K+ +RGY EK+ ++++G P I + T
Sbjct: 338 TRRFFELPYEEKAKIKMTPANGFRGY-----EKIG-ENITEGAPDMHEAIDCYREVTKGM 499
Query: 110 --------VRLNQWPSN-GVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSL 160
NQWP N F F ++ Y + K K+++ IAL+L
Sbjct: 500 YGDLGKVMEGFNQWPQNPPKFKFL----MEEYISL---------CKDLAKKIMRGIALAL 640
Query: 161 NLDENYFEKISALNKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLL-LTNGVPGLQ 219
FE A P +R++ YP G + + G AH+DYG++TLL + + LQ
Sbjct: 641 GGSPYEFEGDRA-GDPFWVMRIIGYP---GVSSDDIGCGAHTDYGLLTLLNQDDDINALQ 808
Query: 220 ICKEKLKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVM-PTGKERYSVAFFM 278
+ W V G + NIGDM++ ++N LY STLHRV+ + K R SV +F
Sbjct: 809 V----RNLSGEWISAPPVPGTFVCNIGDMLKIYSNGLYESTLHRVINNSSKYRVSVVYFY 976
Query: 279 DPPSDCVVE 287
+ D VE
Sbjct: 977 ETNFDAAVE 1003
>TC79860 homologue to PIR|T06533|T06533 probable gibberellin 20-oxidase -
garden pea, partial (93%)
Length = 1454
Score = 97.8 bits (242), Expect = 5e-21
Identities = 88/307 (28%), Positives = 143/307 (45%), Gaps = 12/307 (3%)
Frame = +3
Query: 7 LSLPIIDLSS-----PDRISTAKS-IHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFS 60
L +P IDL + P IS A S ++ AC ++GFF +VNHGVD+ + QA +FF
Sbjct: 177 LQVPPIDLKAFLSGDPKAISNACSQVNDACKKHGFFLVVNHGVDKKLLAQAHKLVDDFFC 356
Query: 61 LPIEDKIKLNRK--EYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPS- 117
+ + +K K RK E+ GY + + P +T S R + S
Sbjct: 357 MQLCEKQKAQRKVGEHCGYANSFIGRFSSKL-------------PWKETLSFRYSDDKSC 497
Query: 118 NGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKEL--LSLIALSLNLDENYFEKISALNK 175
V +F + +++ Q G + Y S L + L+ +SL +D+ YF N
Sbjct: 498 RTVEDYFVNVMGEDFRQFGS-VYQDYCEAMSNLSLGIMELLGMSLGVDKEYFRHFFEAN- 671
Query: 176 PEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVS 235
++ +RL YP P+ + G H D +T+L + V GLQ+ + +W +
Sbjct: 672 -DSVMRLNYYPPCKNPDLAL-GTGPHCDPTSLTILHQDQVEGLQVLVD-----GIWHSIV 830
Query: 236 HVEGAIIVNIGDMMERWTNCLYRSTLHR-VMPTGKERYSVAFFMDPPSDCVVECFESCCS 294
E A +VNIGD +N +++S LHR V+ R S+AFF+ P + +V + +
Sbjct: 831 PKEDAFVVNIGDTFMALSNGIFKSCLHRAVVNDTIVRKSLAFFLCPNEEKIVTPPKELIN 1010
Query: 295 ESSPPRY 301
+ +P Y
Sbjct: 1011KENPMIY 1031
>TC78062 weakly similar to GP|9280317|dbj|BAB01696.1 oxylase-like protein
{Arabidopsis thaliana}, partial (80%)
Length = 1322
Score = 80.9 bits (198), Expect(2) = 5e-21
Identities = 51/153 (33%), Positives = 81/153 (52%), Gaps = 1/153 (0%)
Frame = +3
Query: 151 ELLSLIALSLNLDENYFEKISALNKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLL 210
+++ LIALSL L F+ +++RL YP P + + G H D G +T+L
Sbjct: 609 KIMELIALSLGLPPKRFDDF--FKDETSWIRLNHYPPCPNP-DIVLGCGRHKDSGALTVL 779
Query: 211 LTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVM-PTGK 269
+ V GL++ + K W V + A I+N+GD+++ W+N Y+S HRVM T K
Sbjct: 780 AQDEVSGLEV---RRKSDGEWVLVKPLPNAYIINVGDVIQVWSNDEYKSVEHRVMLNTEK 950
Query: 270 ERYSVAFFMDPPSDCVVECFESCCSESSPPRYR 302
R S FF+ P +VE + ++ +PP+YR
Sbjct: 951 ARLSYPFFLFPSHYTMVEPLKELTNDQNPPKYR 1049
Score = 37.4 bits (85), Expect(2) = 5e-21
Identities = 38/157 (24%), Positives = 62/157 (39%), Gaps = 30/157 (19%)
Frame = +2
Query: 8 SLPIIDLS----------SPDRIST-AKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSA 56
++P+IDLS +P I + I AC E+GFF ++NHG + ++
Sbjct: 158 NIPVIDLSPILDDSNNVKNPLVIDELVRKIGSACKEWGFFQVINHGAPLESRQRIESVGK 337
Query: 57 NFFSLPIEDKIKLNR--------------KEYRGYTPLY-----AEKLDPSSLSKGDPKE 97
FF +E+K K+ R K R + ++ L P+S+ D +
Sbjct: 338 KFFGQKMEEKKKVRRDIVNVMGYYETEHTKNVRDWKEVFDFTVKEPTLVPASIDPDDKEV 517
Query: 98 SYYIGPLTDTTSVRLNQWPSNGVFSFFPAMTLQNYSQ 134
+++ NQWP F T Q Y+Q
Sbjct: 518 THW-----------YNQWPE---FPLEMRETFQEYAQ 586
>TC88003 weakly similar to PIR|T04184|T04184 hypothetical protein F7L13.70 -
Arabidopsis thaliana, partial (26%)
Length = 1209
Score = 95.9 bits (237), Expect = 2e-20
Identities = 82/307 (26%), Positives = 132/307 (42%), Gaps = 12/307 (3%)
Frame = +2
Query: 8 SLPIIDLS-----SPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLP 62
S+PI DLS +P + I +AC E+GFF +VNHGV + + NFF L
Sbjct: 125 SIPISDLSYCDGNNPSSLEVIHKISKACEEFGFFQIVNHGVPDQVCTKMMKAITNFFELA 304
Query: 63 IEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFS 122
E++ L+ + L+ L K + P +P + +
Sbjct: 305 PEEREHLSSTDNTKNVRLFNYYLQVDGGEKVKLWSECFAHP----------WYPIDDIIQ 454
Query: 123 FFPAMTLQNYSQIGDPQWNPYFGKYSG------KELLSLIALSLNLDENYFEKISALNKP 176
P +IG Q+ F +Y+ + LLSLI++ L L+E+ K +P
Sbjct: 455 LLP-------EKIG-TQYREAFTEYAKEVGSLVRRLLSLISIGLGLEEDCLLK-KLGEQP 607
Query: 177 EAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSH 236
+ YP P E G + H+D +T+LL + V GLQ+ K+ W +
Sbjct: 608 RQRAQSNFYPPCPDP-ELTMGLNEHTDLNALTVLLQSEVSGLQVNKD-----GKWISIPC 769
Query: 237 VEGAIIVNIGDMMERWTNCLYRSTLHRVMPTG-KERYSVAFFMDPPSDCVVECFESCCSE 295
+ A ++N+ D +E +N Y+S LHR + + R S+A F P + ++ E
Sbjct: 770 IPNAFVINLADQIEVLSNGRYKSVLHRAVTNNVQPRISMAMFYGPNPETIIGPIHELIDE 949
Query: 296 SSPPRYR 302
PP+YR
Sbjct: 950 EHPPKYR 970
>TC89553 weakly similar to SP|P24397|HY6H_HYONI Hyoscyamine 6-dioxygenase
(EC 1.14.11.11) (Hyoscyamine 6-beta- hydroxylase).
[Henbane], partial (49%)
Length = 1119
Score = 95.9 bits (237), Expect = 2e-20
Identities = 87/307 (28%), Positives = 141/307 (45%), Gaps = 10/307 (3%)
Frame = +1
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
++P+IDL D T I +A EYGFF ++NHGV +D + +AF+ F ++P ++KI
Sbjct: 127 TIPLIDLGGHDHAHTILQILKASEEYGFFQVINHGVSKDLMDEAFNIFQEFHAMPPKEKI 306
Query: 68 KLNRKEYRGYT-PLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVF-SFFP 125
++ G +YA E+Y I + + P +G F F+P
Sbjct: 307 SECSRDPNGINCKIYAS------------SENYKIDAVQYWKDTLTHPCPPSGEFMEFWP 450
Query: 126 AMTLQNYSQIGDPQWNPYFGKYS------GKELLSLIALSLNLDENYFEKISALNKPEAF 179
P++ GKY+ G E+L ++ L L YF I L++
Sbjct: 451 QK---------PPKYREVVGKYTQELNKLGHEILEMLCEGLGLKLEYF--IGELSENPVI 597
Query: 180 LRLLRYPGELGPNEEICGASAHSDYGMITLLLTN-GVPGLQICKEKLKQPQVWEDVSHVE 238
L +P P+ + G + H D +IT+LL + V GLQI K+ W V +
Sbjct: 598 LG-HHFPPCPEPSLTL-GLAKHRDPTIITILLQDQEVHGLQILKD-----DEWIPVEPIP 756
Query: 239 GAIIVNIGDMMERWTNCLYRSTLHRVMPTGKE-RYSVAFFMDPPSDCVVECFESCCSESS 297
A++VNIG +++ TN HRV+ K R SVA+F+ P ++E + ++
Sbjct: 757 NALVVNIGLILQIITNGRLVGAEHRVVTNSKSVRTSVAYFIYPSFSRMIEPAKELVDGNN 936
Query: 298 PPRYRKV 304
PP Y+ +
Sbjct: 937 PPIYKSM 957
>TC88118 weakly similar to PIR|T09683|T09683 gibberellin 7-oxidase (EC
1.14.11.-) - winter squash, partial (68%)
Length = 1220
Score = 94.0 bits (232), Expect = 7e-20
Identities = 88/308 (28%), Positives = 137/308 (43%), Gaps = 11/308 (3%)
Frame = +2
Query: 9 LPIIDLS-------SPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSL 61
+P IDLS ++ ++I +AC EYGFF +VNHG+ D +KQA + S FF+
Sbjct: 107 IPTIDLSPFLKENKDDNKKKAMETITKACSEYGFFQIVNHGISLDLMKQAMELSKTFFNY 286
Query: 62 PIEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVF 121
E+K N+ PL A S S P ++ Y L +P F
Sbjct: 287 SDEEK---NKSSPSSNAPLPAGYSRQPSHS---PDKNEY-----------LLVFPPRSNF 415
Query: 122 SFFPAMTLQNYSQIGDPQWNPYFGKYS--GKELLSLIALSLNLDENYFEKISALNKPEAF 179
+ +P +N Q+ + F + S G + ++I L L N+ ++ + +
Sbjct: 416 NVYP----ENPPQLREVV-EELFAQTSKIGVVIENIINECLGLPPNFLKEFNNDRSWDIM 580
Query: 180 LRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEG 239
+ P N E G H D +TL+L + V GL++ + W V EG
Sbjct: 581 VAFRYIPAS---NNENVGLREHQDVNCVTLVLQDEVGGLEVLSN-----EEWVPVVPAEG 736
Query: 240 AIIVNIGDMMERWTNCLYRSTLHRVMPTG-KERYSVAFFMDPPSDCVVECFESCCSE-SS 297
I+VN+GD+++ +N ++S HRV+ G K RYS AFF + + VE E
Sbjct: 737 TIVVNVGDVIQVLSNKKFKSASHRVVRKGEKSRYSFAFFHNLNGEKWVEPLPQFTKEIGE 916
Query: 298 PPRYRKVL 305
P+YR L
Sbjct: 917 SPKYRGFL 940
>TC77423 weakly similar to PIR|D96569|D96569 probable oxidoreductase
38288-39393 [imported] - Arabidopsis thaliana, partial
(87%)
Length = 1141
Score = 91.3 bits (225), Expect = 4e-19
Identities = 88/311 (28%), Positives = 129/311 (41%), Gaps = 9/311 (2%)
Frame = +1
Query: 1 MAGTATLSLPIIDLS-------SPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFD 53
M AT LPIID + SP+ +++A VEYG F V V D K F
Sbjct: 25 MGTEATPKLPIIDFNNLNLEAKSPNWELVKSQVYKALVEYGCFEAVFDKVPLDLRKAIFA 204
Query: 54 QSANFFSLPIEDK-IKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRL 112
F LP+E K + +++K Y GY Y S+ D + +T+
Sbjct: 205 SLQELFDLPLETKLLNVSKKPYHGYVGQYPMIPLFESMGIDDANILEKVDSMTNNLWPDG 384
Query: 113 NQWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISA 172
NQ S + SF +T QI + +I SL + E Y E+
Sbjct: 385 NQNFSKTIHSFSEELT--ELDQI----------------IRKMILESLGV-EKYLEE--H 501
Query: 173 LNKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWE 232
+N LR+++Y G + ++ G S HSD ++T+L N V GL++ K Q
Sbjct: 502 MNSTNYLLRVMKYKGPQTSDTKL-GLSTHSDKNVVTILYQNQVEGLEV---MTKDGQWIS 669
Query: 233 DVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKE-RYSVAFFMDPPSDCVVECFES 291
G+ + IGD + W+N S HRVM +G E RYS F P +++ E
Sbjct: 670 YKPSSSGSFVAMIGDSIHAWSNGRLHSPFHRVMMSGNEARYSTGLFSIPKGGYIIKAPEE 849
Query: 292 CCSESSPPRYR 302
E P +R
Sbjct: 850 LVDEEHPLLFR 882
>TC90278 weakly similar to GP|9280318|dbj|BAB01697.1 oxidase-like protein
{Arabidopsis thaliana}, partial (21%)
Length = 1139
Score = 66.6 bits (161), Expect(2) = 5e-18
Identities = 41/123 (33%), Positives = 64/123 (51%), Gaps = 1/123 (0%)
Frame = +2
Query: 181 RLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGA 240
R+ YP P E G H D G +T+L + V GL++ K + Q W V A
Sbjct: 656 RINHYPPCPSP-ELALGMGRHKDAGALTILAQDEVAGLEV---KHQISQEWVLVKPTPNA 823
Query: 241 IIVNIGDMMERWTNCLYRSTLHRVMPT-GKERYSVAFFMDPPSDCVVECFESCCSESSPP 299
I+N+GD+ + W+N Y S HRV+ KER+S+AF + P V+ + ++ +PP
Sbjct: 824 FIINVGDLTQVWSNDEYESVEHRVIVNFEKERFSIAFGVFPAHYIEVKPLDELINDENPP 1003
Query: 300 RYR 302
+Y+
Sbjct: 1004KYK 1012
Score = 41.6 bits (96), Expect(2) = 5e-18
Identities = 49/166 (29%), Positives = 66/166 (39%), Gaps = 23/166 (13%)
Frame = +1
Query: 21 STAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSAN-FFSLPIEDKIKLNRKEYRGYTP 79
S K I AC E+GFF + NH V I+Q D++A FF+ +E+K+K+ R E TP
Sbjct: 193 SLVKKIGSACKEWGFFQVTNHEVPLS-IRQRLDEAARLFFAQSLEEKMKVARDE---ITP 360
Query: 80 LYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFF---PAMTLQNYSQIG 136
IG + + W VF FF P + N +
Sbjct: 361 ---------------------IGYFDKEHTKNVRNWIE--VFDFFARDPTLVPLNCDEND 471
Query: 137 DP--QWNPYFGKYSGK-----------------ELLSLIALSLNLD 163
D QW+ KY K +LL LIALSL L+
Sbjct: 472 DQVIQWSNPSPKYPPKFRNIIEEYIKEMENLASKLLELIALSLGLE 609
>TC85507 homologue to SP|P31239|ACCO_PEA 1-aminocyclopropane-1-carboxylate
oxidase (EC 1.-.-.-) (ACC oxidase) (Ethylene-forming
enzyme) (EFE), complete
Length = 1342
Score = 87.0 bits (214), Expect = 8e-18
Identities = 82/291 (28%), Positives = 135/291 (46%), Gaps = 12/291 (4%)
Frame = +1
Query: 8 SLPIID---LSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIE 64
+ P++D L++ +R +T + I AC +GFF VNH + S+ +
Sbjct: 103 NFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSI----------------SIELM 234
Query: 65 DKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFF 124
DK++ KE+ Y ++ SKG I L ++ L PS+ + S
Sbjct: 235 DKVEKLTKEH--YKKCMEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNI-SEI 405
Query: 125 PAMTLQNYSQIGDPQWNPYFGKYSGK------ELLSLIALSLNLDENYFEKISALNK-PE 177
P + D + +++ K ELL L+ +L L++ Y +K+ +K P
Sbjct: 406 PDL---------DEDYRKTMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPN 558
Query: 178 AFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNG-VPGLQICKEKLKQPQVWEDVSH 236
++ YP P + I G AH+D G I LL + V GLQ+ K+ W DV
Sbjct: 559 FGTKVSNYPPCPKP-DLIKGLRAHTDAGGIILLFQDDKVSGLQLLKD-----DQWIDVPP 720
Query: 237 VEGAIIVNIGDMMERWTNCLYRSTLHRVM-PTGKERYSVAFFMDPPSDCVV 286
+ +I++N+GD +E TN Y+S +HRV+ T R S+A F +P +D V+
Sbjct: 721 MPHSIVINLGDQLEVITNGKYKSVMHRVIAQTDGARMSIASFYNPGNDAVI 873
>BQ122436 weakly similar to PIR|G86472|G86 probable hyoscyamine 6-dioxygenase
hydroxylase [imported] - Arabidopsis thaliana, partial
(40%)
Length = 658
Score = 87.0 bits (214), Expect = 8e-18
Identities = 42/93 (45%), Positives = 61/93 (65%), Gaps = 1/93 (1%)
Frame = +3
Query: 6 TLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIED 65
T L IDLS+P+ + Q+C + GFFYL+NHG+ +DF+ + F QS FF+LP+ +
Sbjct: 27 TAILNCIDLSNPNIDHNVDLLKQSCTDSGFFYLINHGISQDFMDEVFAQSNTFFTLPLNE 206
Query: 66 KIKLNRKE-YRGYTPLYAEKLDPSSLSKGDPKE 97
K+KL R + ++GYTP+ E LDP + GD KE
Sbjct: 207 KMKLLRNDSHKGYTPVLDEILDPQNQLHGDYKE 305
Score = 29.6 bits (65), Expect = 1.5
Identities = 13/24 (54%), Positives = 16/24 (66%)
Frame = +2
Query: 162 LDENYFEKISALNKPEAFLRLLRY 185
LD N+F+K L +P A LRLL Y
Sbjct: 572 LDANFFDKPETLREPIAILRLLHY 643
>TC78764 similar to PIR|C96802|C96802 hypothetical protein F2P24.4
[imported] - Arabidopsis thaliana, partial (88%)
Length = 1276
Score = 86.7 bits (213), Expect = 1e-17
Identities = 86/291 (29%), Positives = 133/291 (45%), Gaps = 16/291 (5%)
Frame = +1
Query: 7 LSLPIID---LSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPI 63
+++P+ID L+ +R T I C E+GFF L+NHG+ E+ +++ S+ F+ L
Sbjct: 67 MAVPVIDFSKLNGEERAKTLAQIANGCEEWGFFQLINHGISEELLERVKKVSSEFYKLER 246
Query: 64 EDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSF 123
E+ K N K + + AEK L D ++ I L D N+WP N
Sbjct: 247 EENFK-NSKTVKLLNDI-AEKKSSEKLENVDWED--VITLLDD------NEWPEN----- 381
Query: 124 FPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDEN------YFEKISALNKPE 177
P + +Y + ++L+ +DEN Y +K ALN E
Sbjct: 382 -------------TPSFRETMSEYRSELKKLAVSLTEVMDENLGLPKGYIKK--ALNDGE 516
Query: 178 ---AFL--RLLRYPGELGPNEEICGASAHSDYGMITLLLTNG-VPGLQICKEKLKQPQVW 231
AF ++ YP P E + G AH+D G + LL + V GLQ+ K+ W
Sbjct: 517 GDNAFFGTKVSHYPPCPHP-ELVNGLRAHTDAGGVILLFQDDKVGGLQMLKD-----GEW 678
Query: 232 EDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMP-TGKERYSVAFFMDPP 281
DV + AI++N GD +E +N +S HRV+ T R ++A F +PP
Sbjct: 679 LDVQPLPNAIVINTGDQIEVLSNGRNKSCWHRVLNFTEGTRRTIASFYNPP 831
>AW690062 weakly similar to GP|7268917|emb gibberellin 20-oxidase-like
protein {Arabidopsis thaliana}, partial (44%)
Length = 629
Score = 82.8 bits (203), Expect = 2e-16
Identities = 55/155 (35%), Positives = 88/155 (56%), Gaps = 1/155 (0%)
Frame = +1
Query: 150 KELLSLIALSLNLDENYFEKISALNKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITL 209
+ L+ ++A LN++ +YF++ + N ++LRL RYP P++ I G H+D IT+
Sbjct: 133 ENLVQILAQELNINFSYFQQNCSANT--SYLRLNRYPPCPFPSKVI-GLLPHADTSFITI 303
Query: 210 LLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGK 269
+ + + GLQ+ K+ W V A+IVN+GD+ + +N LY S HRV+ K
Sbjct: 304 VHQDHIGGLQLMKDGK-----WISVKPNSEALIVNVGDLFQALSNGLYTSVGHRVVAAEK 468
Query: 270 -ERYSVAFFMDPPSDCVVECFESCCSESSPPRYRK 303
ER+S+A+F P D V+E S ++PP YRK
Sbjct: 469 VERFSLAYFYGPSIDAVIE------SYATPPLYRK 555
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,782,600
Number of Sequences: 36976
Number of extensions: 163482
Number of successful extensions: 965
Number of sequences better than 10.0: 167
Number of HSP's better than 10.0 without gapping: 851
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 888
length of query: 309
length of database: 9,014,727
effective HSP length: 96
effective length of query: 213
effective length of database: 5,465,031
effective search space: 1164051603
effective search space used: 1164051603
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149079.14


