

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
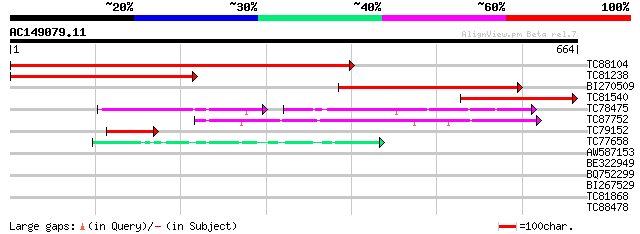
Query= AC149079.11 - phase: 0
(664 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88104 similar to GP|15553480|gb|AAL01888.1 acyl-CoA oxidase {G... 813 0.0
TC81238 homologue to GP|15553478|gb|AAL01887.1 acyl-CoA oxidase ... 401 e-112
BI270509 similar to GP|15553478|gb acyl-CoA oxidase {Glycine max... 369 e-102
TC81540 homologue to GP|15553480|gb|AAL01888.1 acyl-CoA oxidase ... 281 6e-76
TC78475 similar to GP|20466227|gb|AAM20431.1 acyl-CoA oxidase AC... 138 3e-53
TC87752 similar to GP|10178173|dbj|BAB11647. acyl-CoA oxidase {A... 195 4e-50
TC79152 similar to PIR|T07901|T07901 acyl CoA oxidase homolog - ... 69 8e-12
TC77658 similar to PIR|T46895|T46895 acyl-CoA dehydrogenase (EC ... 65 8e-11
AW587153 PIR|T52121|T52 acyl-CoA oxidase (EC 1.3.3.6) peroxisom... 41 0.002
BE322949 similar to GP|7293851|gb| CG3902-PA {Drosophila melanog... 33 0.27
BQ752299 similar to GP|11595639|e conserved hypothetical protein... 29 5.1
BI267529 similar to GP|18460924|gb sialoglycoprotease GCP1 {Arab... 29 6.6
TC81868 homologue to PIR|T09538|T09538 acetyl-CoA carboxylase (E... 29 6.6
TC88478 weakly similar to GP|20258927|gb|AAM14179.1 unknown prot... 28 8.7
>TC88104 similar to GP|15553480|gb|AAL01888.1 acyl-CoA oxidase {Glycine
max}, partial (60%)
Length = 1345
Score = 813 bits (2099), Expect = 0.0
Identities = 400/404 (99%), Positives = 401/404 (99%)
Frame = +1
Query: 1 MEGEDHLAFERNKAEFDVDAMKIVWAGSAHALEVSDRMSRLVASDPAFRKDHRPMLGRKE 60
MEGEDHLAFERNKAEFDVDAMKIVWAGSAHALEVSDRMSRLVASDPAFRKDHRPMLGRKE
Sbjct: 133 MEGEDHLAFERNKAEFDVDAMKIVWAGSAHALEVSDRMSRLVASDPAFRKDHRPMLGRKE 312
Query: 61 LFKSTLKKAAYAWKRIIELRLTEEEASMLRSFVDEPAFTDLHWGMFIPAIKGQGTEEQQQ 120
LFKSTLKKAAYAWKRIIELRLTEEEASMLRSFVDEPAFTDLHWGMFIPAIKGQGTEEQQQ
Sbjct: 313 LFKSTLKKAAYAWKRIIELRLTEEEASMLRSFVDEPAFTDLHWGMFIPAIKGQGTEEQQQ 492
Query: 121 KWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSKWWPGGLG 180
KWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSKWWPGGLG
Sbjct: 493 KWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSKWWPGGLG 672
Query: 181 KVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGITVGDIGMKFGNGAYNTMDNG 240
KVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGITVGDIGMKFGNGAYNTMDNG
Sbjct: 673 KVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGITVGDIGMKFGNGAYNTMDNG 852
Query: 241 VLRFEHVRIPRDQMLMRVSQVTREGKYVQSNVPRQLVYGTMVYVRQTIVSDASTAMSRAV 300
VLRFEHVRIPRDQMLMRVSQVTREGKYVQSNVPRQLVYGTMVYVRQTIVSDASTAMSRAV
Sbjct: 853 VLRFEHVRIPRDQMLMRVSQVTREGKYVQSNVPRQLVYGTMVYVRQTIVSDASTAMSRAV 1032
Query: 301 CIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFPLLASAYAFRFVGEWLKWLYTDVMK 360
CIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFPLLASAYAFR GEW+KWLYTDVMK
Sbjct: 1033CIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFPLLASAYAFRIGGEWVKWLYTDVMK 1212
Query: 361 RLQANDFSTLPEAHACTAGLKSLTTSATSDGIEECRKLCGGHGY 404
RLQANDFSTLPEAHACT GLKSLTTSATSDGIEECRKLCGGHGY
Sbjct: 1213RLQANDFSTLPEAHACTVGLKSLTTSATSDGIEECRKLCGGHGY 1344
>TC81238 homologue to GP|15553478|gb|AAL01887.1 acyl-CoA oxidase {Glycine
max}, partial (32%)
Length = 866
Score = 401 bits (1030), Expect = e-112
Identities = 190/220 (86%), Positives = 206/220 (93%)
Frame = +1
Query: 1 MEGEDHLAFERNKAEFDVDAMKIVWAGSAHALEVSDRMSRLVASDPAFRKDHRPMLGRKE 60
MEG DHLAFERNKAEFDV+ MKIVWAGS E+SDR+SRLVASDPAFRKD+R L RKE
Sbjct: 205 MEGVDHLAFERNKAEFDVNEMKIVWAGSRQEFELSDRISRLVASDPAFRKDNRTSLDRKE 384
Query: 61 LFKSTLKKAAYAWKRIIELRLTEEEASMLRSFVDEPAFTDLHWGMFIPAIKGQGTEEQQQ 120
LFK+TL+K AYAWKRIIELRL E+EAS LRSFVDEPAFTDLHWGMF+PAIKGQGT+EQQ+
Sbjct: 385 LFKNTLRKTAYAWKRIIELRLNEQEASKLRSFVDEPAFTDLHWGMFVPAIKGQGTDEQQE 564
Query: 121 KWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSKWWPGGLG 180
KWLPLA +MQIIGCYAQTELGHGSNVQGLETTATFDPKTDEF+IHSPTLTSSKWWPGGLG
Sbjct: 565 KWLPLAYKMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFIIHSPTLTSSKWWPGGLG 744
Query: 181 KVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGI 220
K+STHA+VYARLIT+G+D GVHGFIVQLRSLDDHLPLPGI
Sbjct: 745 KISTHAIVYARLITEGQDQGVHGFIVQLRSLDDHLPLPGI 864
>BI270509 similar to GP|15553478|gb acyl-CoA oxidase {Glycine max}, partial
(32%)
Length = 646
Score = 369 bits (948), Expect = e-102
Identities = 180/215 (83%), Positives = 201/215 (92%)
Frame = +2
Query: 386 SATSDGIEECRKLCGGHGYLCSSGLPELFAVYVPTCTYEGDNIVLLLQVARHLMKVISRL 445
SAT+DGIEECRKLCGGHGYLCSSGLPELFAVYVP CTYEGDN VLLLQVAR+L+K IS+L
Sbjct: 2 SATADGIEECRKLCGGHGYLCSSGLPELFAVYVPACTYEGDNTVLLLQVARYLVKTISQL 181
Query: 446 GSGKKPVGTTAYMGRVEQLLEARSDVQKAEDWLNPNTVLRAFEARAARMSVACAKNLTKF 505
GSGKKPVGTTAY+GRVEQL++ +SDV++AEDWL PN V+ AFEARAARMSVA A+N++KF
Sbjct: 182 GSGKKPVGTTAYLGRVEQLMQYQSDVKRAEDWLKPNVVIEAFEARAARMSVAVAQNISKF 361
Query: 506 SNPEEGFQELLADLVDAARAHCQLIVVSKFIEKLQQDIPGKGVKRQLEILCNIYALFHLH 565
SNPEEGFQEL DLV+AA AHCQLIVVSKFIEKLQQ+IPGKGVK+QLE+LC+IYALF LH
Sbjct: 362 SNPEEGFQELSVDLVEAAAAHCQLIVVSKFIEKLQQNIPGKGVKQQLEVLCSIYALFLLH 541
Query: 566 KHLGDFLSTGCITPKQGSLANDQLRSLYSQVRPNA 600
KHLGDFLSTG IT +QGSLAN+QLRSLYSQVRPNA
Sbjct: 542 KHLGDFLSTGSITEEQGSLANEQLRSLYSQVRPNA 646
>TC81540 homologue to GP|15553480|gb|AAL01888.1 acyl-CoA oxidase {Glycine
max}, partial (20%)
Length = 926
Score = 281 bits (719), Expect = 6e-76
Identities = 137/137 (100%), Positives = 137/137 (100%)
Frame = +1
Query: 528 QLIVVSKFIEKLQQDIPGKGVKRQLEILCNIYALFHLHKHLGDFLSTGCITPKQGSLAND 587
QLIVVSKFIEKLQQDIPGKGVKRQLEILCNIYALFHLHKHLGDFLSTGCITPKQGSLAND
Sbjct: 1 QLIVVSKFIEKLQQDIPGKGVKRQLEILCNIYALFHLHKHLGDFLSTGCITPKQGSLAND 180
Query: 588 QLRSLYSQVRPNAIALVDAFNYTDHYLSSVLGRYDGNVYPKLYEEAWKDPLNDSVVPDGF 647
QLRSLYSQVRPNAIALVDAFNYTDHYLSSVLGRYDGNVYPKLYEEAWKDPLNDSVVPDGF
Sbjct: 181 QLRSLYSQVRPNAIALVDAFNYTDHYLSSVLGRYDGNVYPKLYEEAWKDPLNDSVVPDGF 360
Query: 648 QEYIRPMLKQQLRNARL 664
QEYIRPMLKQQLRNARL
Sbjct: 361 QEYIRPMLKQQLRNARL 411
>TC78475 similar to GP|20466227|gb|AAM20431.1 acyl-CoA oxidase ACX3
{Arabidopsis thaliana}, partial (90%)
Length = 2177
Score = 138 bits (348), Expect(2) = 3e-53
Identities = 77/210 (36%), Positives = 123/210 (57%), Gaps = 10/210 (4%)
Frame = +3
Query: 103 WGMFIPAIKGQGTEEQQQKWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEF 162
WG A+K GT+ KWL + I GC+A +ELGHGSNV+G+ET T+DP + EF
Sbjct: 519 WG---GAVKFLGTKRHHDKWLRATENYDIKGCFAMSELGHGSNVRGIETVTTYDPISGEF 689
Query: 163 VIHSPTLTSSKWWPGGLGKVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGITV 222
VI++P ++ K+W GG +TH +V+++L +G + GVH FI Q+R+ D ++ P I +
Sbjct: 690 VINTPCESAQKYWIGGAANHATHTIVFSQLNINGSNQGVHAFIAQIRNSDGNI-CPNIRI 866
Query: 223 GDIGMKFGNGAYNTMDNGVLRFEHVRIPRDQMLMRVSQVTREGKYVQS--NVPRQ----- 275
+ G K G N +DNG + F++VRIPR+ + ++ V+ G+Y+ + N ++
Sbjct: 867 AECGHKIG---LNGVDNGRIWFDNVRIPRENL*NSMADVSPSGEYLSAIKNTDQRFAAFL 1037
Query: 276 --LVYGTM-VYVRQTIVSDASTAMSRAVCI 302
L G + + V +S S A+S +CI
Sbjct: 1038APLTSGRVTIAVSSVYISKISLAISYKICI 1127
Score = 89.4 bits (220), Expect(2) = 3e-53
Identities = 77/301 (25%), Positives = 140/301 (45%), Gaps = 4/301 (1%)
Frame = +1
Query: 321 ESQVIDYKTQQARLFPLLASAYAFRFVGEWLKWLYTDVMKRLQANDFSTLPEAHACTAGL 380
E ++DY + Q RL PLLA YA F LK LY V + ++N H ++
Sbjct: 1168 EVLLLDYPSHQQRLLPLLAKIYAMSFSAIELKKLY--VNRTPESNK-----AIHIVSSAY 1326
Query: 381 KSLTTSATSDGIEECRKLCGGHGYLCSSGLPELFAVYVPTCTYEGDNIVLLLQVARHLMK 440
K+ T ++ECR+ CGG G + + + Y T+EGDN VL+ Q+++ L+
Sbjct: 1327 KATFTWNNMRTLQECREACGGQGVKTENRIGQFKGEYDVHSTFEGDNNVLMQQISKALLA 1506
Query: 441 VISRLGSGKKP---VGTTAYMGRVEQL-LEARSDVQKAEDWLNPNTVLRAFEARAARMSV 496
+ KP +G G + + S K+ ++ LR + R +
Sbjct: 1507 EYIACQTKNKPFKGLGLEHMNGSCPVIPSQLTSSTIKSSEFQIDLFHLRERDL-LKRFAA 1683
Query: 497 ACAKNLTKFSNPEEGFQELLADLVDAARAHCQLIVVSKFIEKLQQDIPGKGVKRQLEILC 556
+++ ++ ++ E F D RA + ++ F+E + +P +K L +L
Sbjct: 1684 EVSEHQSQGNSKESAFILSYQLAEDLGRAFSERAILKTFME-AESALPAGSLKDVLRLLR 1860
Query: 557 NIYALFHLHKHLGDFLSTGCITPKQGSLANDQLRSLYSQVRPNAIALVDAFNYTDHYLSS 616
++YAL + + FL G ++ + S ++ L +++RP+A+ALV++F D +LS
Sbjct: 1861 SLYALISVDEDAA-FLRYGYLSTENASAVRKEVPKLCAELRPHALALVNSFGIPDAFLSP 2037
Query: 617 V 617
+
Sbjct: 2038 I 2040
>TC87752 similar to GP|10178173|dbj|BAB11647. acyl-CoA oxidase {Arabidopsis
thaliana}, partial (61%)
Length = 1524
Score = 195 bits (496), Expect = 4e-50
Identities = 140/419 (33%), Positives = 208/419 (49%), Gaps = 12/419 (2%)
Frame = +3
Query: 217 LPGITVGDIGMKFGNGAYNTMDNGVLRFEHVRIPRDQMLMRVSQVTREGKYVQS----NV 272
LPGI + D G K G N +DNG LRF VRIPRD +L R V+R+GKY + N
Sbjct: 9 LPGIEIHDCGHKVG---LNGVDNGALRFRSVRIPRDNLLNRFGDVSRDGKYTSTLPSVNK 179
Query: 273 PRQLVYGTMVYVRQTIVSDASTAMSRAVCIAARYSAVRRQFGGNNGSLESQVIDYKTQQA 332
G +V R + + + + + IA RYS +R+QFG + E ++DY++ Q
Sbjct: 180 RFGATLGELVGGRVGLAYSSVSVLKVSATIAIRYSLLRQQFGPRD-QPEVTILDYQSHQH 356
Query: 333 RLFPLLASAYAFRFVGEWLKWLYTDVMKRLQANDFSTLPEAHACTAGLKSLTTSATSDGI 392
+L P+LAS YAF F L Y + K ++D + + HA +AGLK+ TS T+ +
Sbjct: 357 KLMPMLASTYAFHFATTNLVEKYAQMKK---SHDEELVADVHALSAGLKAYVTSYTAKSL 527
Query: 393 EECRKLCGGHGYLCSSGLPELFAVYVPTCTYEGDNIVLLLQVARHLMKVISRLGSGKKPV 452
CR+ CGGHGY + L + T+EGDN VLL QVA L+K R G
Sbjct: 528 SICREACGGHGYAAVNRFGSLRNDHDIFQTFEGDNTVLLQQVAADLLKQYQRKFQGGTLA 707
Query: 453 GTTAYM-GRVEQLLEARSDVQ---KAEDWL-NPNTVLRAFEARAARMSVACAKNLTKFSN 507
T Y+ + L + V + ED L +P L AF R +R+ + A L K S
Sbjct: 708 VTWNYLRDSMNTYLSQPNPVTARWEGEDHLRDPKFQLDAFRYRTSRLLHSVAVRLRKHSK 887
Query: 508 PEEGF---QELLADLVDAARAHCQLIVVSKFIEKLQQDIPGKGVKRQLEILCNIYALFHL 564
P F L L+ A +H + ++++KFIE + Q P + L+++C++YAL +
Sbjct: 888 PLGDFGAWNRCLNHLLTLAESHIESVILAKFIEAV-QSCPDASAQAALKLVCDLYALDRI 1064
Query: 565 HKHLGDFLSTGCITPKQGSLANDQLRSLYSQVRPNAIALVDAFNYTDHYLSSVLGRYDG 623
+G + + + P + + L QVR A LVDAF+ DH + + + G
Sbjct: 1065WNDIGTYRNVDYVAPNKAKAIHKLSEYLSFQVRNIAKELVDAFDLPDHVTRAPIAKQSG 1241
>TC79152 similar to PIR|T07901|T07901 acyl CoA oxidase homolog - cucurbit,
partial (28%)
Length = 873
Score = 68.6 bits (166), Expect = 8e-12
Identities = 31/61 (50%), Positives = 41/61 (66%)
Frame = +2
Query: 114 GTEEQQQKWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSK 173
GT++ + K+ + GC+A TEL HGSNVQGL+T ATFDP TDEF+I +P + K
Sbjct: 650 GTQKHKDKYYDGIDNLDYPGCFAMTELHHGSNVQGLQTLATFDPITDEFIIDTPNDGAIK 829
Query: 174 W 174
W
Sbjct: 830 W 832
>TC77658 similar to PIR|T46895|T46895 acyl-CoA dehydrogenase (EC 1.3.99.3)
[validated] - Arabidopsis thaliana, partial (94%)
Length = 2886
Score = 65.1 bits (157), Expect = 8e-11
Identities = 85/341 (24%), Positives = 137/341 (39%)
Frame = +1
Query: 98 FTDLHWGMFIPAIKGQGTEEQQQKWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDP 157
F +H + + I G+E Q+QK+LP +++ I C+A TE +GS+ L+TTAT
Sbjct: 532 FILVHSSLAMLTIALCGSEAQKQKYLPSLAQLKTIACWALTEPDYGSDASALKTTAT--- 702
Query: 158 KTDEFVIHSPTLTSSKWWPGGLGKVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPL 217
K + I L K W G ST A + + + ++G+IV+ +
Sbjct: 703 KVEGGWI----LEGQKRWIGN----STFADLLVIFARNTSTNQINGYIVKKDA------- 837
Query: 218 PGITVGDIGMKFGNGAYNTMDNGVLRFEHVRIPRDQMLMRVSQVTREGKYVQSNVPRQLV 277
PG+TV I K G + NG + V +P + + V+ K + V R +V
Sbjct: 838 PGLTVTKIENKIG---LRIVQNGDIVMRKVFVPDEDRIEGVNSFQDTNKVLA--VSRVMV 1002
Query: 278 YGTMVYVRQTIVSDASTAMSRAVCIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFPL 337
+ + I + RY R+QFG + + ++ Q +L +
Sbjct: 1003AWQPIGISMGIYD-----------MCHRYLKERKQFG-------APLAAFQINQQKLVQM 1128
Query: 338 LASAYAFRFVGEWLKWLYTDVMKRLQANDFSTLPEAHACTAGLKSLTTSATSDGIEECRK 397
L++ A VG L LY + + HA KS T + R+
Sbjct: 1129LSNVQAMILVGWRLCKLY----------ESGKMTPGHASLG--KSWITLRARETAALGRE 1272
Query: 398 LCGGHGYLCSSGLPELFAVYVPTCTYEGDNIVLLLQVARHL 438
L GG+G L + + F P TYEG + L R +
Sbjct: 1273LLGGNGILADFLVAKAFCDLEPIYTYEGTYEINTLVTGREV 1395
>AW587153 PIR|T52121|T52 acyl-CoA oxidase (EC 1.3.3.6) peroxisomal
[validated] - Arabidopsis thaliana, partial (3%)
Length = 359
Score = 40.8 bits (94), Expect = 0.002
Identities = 36/106 (33%), Positives = 52/106 (48%)
Frame = +1
Query: 1 MEGEDHLAFERNKAEFDVDAMKIVWAGSAHALEVSDRMSRLVASDPAFRKDHRPMLGRKE 60
MEG DHL + K + MKIVWAGS D S ++ S + P L +
Sbjct: 61 MEGVDHLLSKGTKLN-SWNEMKIVWAGSRQN-STFDEFSFVLRS--GIPRITEPRLIERS 228
Query: 61 LFKSTLKKAAYAWKRIIELRLTEEEASMLRSFVDEPAFTDLHWGMF 106
+ +K ++AWK IELRL E++ + + +P F +WGMF
Sbjct: 229 -YLEHFEKNSFAWKD-IELRLNEQKLQL--NLCRQPVFRS-YWGMF 351
>BE322949 similar to GP|7293851|gb| CG3902-PA {Drosophila melanogaster},
partial (28%)
Length = 679
Score = 33.5 bits (75), Expect = 0.27
Identities = 31/99 (31%), Positives = 45/99 (45%)
Frame = +1
Query: 77 IELRLTEEEASMLRSFVDEPAFTDLHWGMFIPAIKGQGTEEQQQKWLPLAQRMQIIGCYA 136
+ + L EE S + V A+ D+H + GTEEQ+QK+L + G +
Sbjct: 370 LTMMLVVEELSRVDPAV--AAYVDIHNTLVNSLFMKLGTEEQKQKYL-TKLCTEYAGSFC 540
Query: 137 QTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSKWW 175
TE GS+ L+T A D D +VI + SK W
Sbjct: 541 LTEPSSGSDAFALKTVAKKD--GDHYVI-----SGSKMW 636
>BQ752299 similar to GP|11595639|e conserved hypothetical protein {Neurospora
crassa}, partial (7%)
Length = 1005
Score = 29.3 bits (64), Expect = 5.1
Identities = 16/46 (34%), Positives = 23/46 (49%)
Frame = +3
Query: 450 KPVGTTAYMGRVEQLLEARSDVQKAEDWLNPNTVLRAFEARAARMS 495
+P+G R +L AR D + WL P TV+R +R R+S
Sbjct: 366 RPLGPRGLSNRHMRLPPARVDPRSQ*TWLRPLTVVRTILSRDGRVS 503
>BI267529 similar to GP|18460924|gb sialoglycoprotease GCP1 {Arabidopsis
thaliana}, partial (29%)
Length = 624
Score = 28.9 bits (63), Expect = 6.6
Identities = 15/38 (39%), Positives = 19/38 (49%), Gaps = 6/38 (15%)
Frame = +1
Query: 378 AGLKSLTTSATSDGIEECRKL------CGGHGYLCSSG 409
+G KSL + ++G E CR C HG CSSG
Sbjct: 436 SGFKSLPSCGGAEGSENCRGF*FTNYWCPSHGGSCSSG 549
>TC81868 homologue to PIR|T09538|T09538 acetyl-CoA carboxylase (EC 6.4.1.2)
- alfalfa, partial (14%)
Length = 994
Score = 28.9 bits (63), Expect = 6.6
Identities = 15/37 (40%), Positives = 21/37 (56%)
Frame = +1
Query: 578 TPKQGSLANDQLRSLYSQVRPNAIALVDAFNYTDHYL 614
TPK+ S ND++ L S ALV F+++DH L
Sbjct: 163 TPKRKSAINDRMEDLVSAPLAVEDALVGLFDHSDHTL 273
>TC88478 weakly similar to GP|20258927|gb|AAM14179.1 unknown protein
{Arabidopsis thaliana}, partial (8%)
Length = 1147
Score = 28.5 bits (62), Expect = 8.7
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +3
Query: 141 GHGSNVQGLETTATFDPKTDEFVIHSP 167
GH N L+T +DPK DE+ + +P
Sbjct: 633 GHDENKNALKTAWAYDPKIDEWTMLAP 713
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,388,053
Number of Sequences: 36976
Number of extensions: 238913
Number of successful extensions: 1092
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1075
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1083
length of query: 664
length of database: 9,014,727
effective HSP length: 102
effective length of query: 562
effective length of database: 5,243,175
effective search space: 2946664350
effective search space used: 2946664350
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149079.11


