

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149038.15 - phase: 0
(213 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
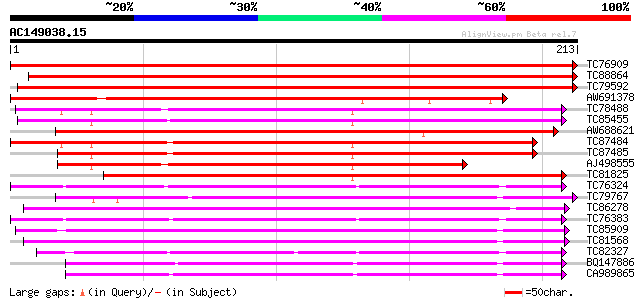
Score E
Sequences producing significant alignments: (bits) Value
TC76909 similar to SP|Q9S8P4|RHRE_PEA Rhicadhesin receptor precu... 416 e-117
TC88864 similar to GP|1934730|gb|AAB51752.1| germin-like protein... 319 6e-88
TC79592 similar to GP|1934730|gb|AAB51752.1| germin-like protein... 275 1e-74
AW691378 similar to SP|Q9S8P4|RHRE Rhicadhesin receptor precurso... 236 6e-63
TC78488 similar to GP|6689036|emb|CAB65370.1 germin-like protein... 177 3e-45
TC85455 similar to GP|6689036|emb|CAB65370.1 germin-like protein... 176 6e-45
AW688621 weakly similar to SP|P92998|GL11 Germin-like protein su... 172 7e-44
TC87484 homologue to GP|6689036|emb|CAB65370.1 germin-like prote... 170 3e-43
TC87485 homologue to GP|6689036|emb|CAB65370.1 germin-like prote... 169 7e-43
AJ498555 weakly similar to SP|Q9FMA8|GL1B Germin-like protein su... 146 5e-36
TC81825 weakly similar to SP|Q9FIC8|GL1G_ARATH Germin-like prote... 146 7e-36
TC76324 similar to GP|7242813|emb|CAB77393.1 germin-like protein... 143 4e-35
TC79767 similar to SP|Q9SR72|GL32_ARATH Germin-like protein subf... 142 7e-35
TC86278 similar to GP|17473703|gb|AAL38307.1 germin-like protein... 141 2e-34
TC76383 similar to GP|7242813|emb|CAB77393.1 germin-like protein... 139 6e-34
TC85909 similar to GP|13277342|emb|CAC34417. Germin-like protein... 133 6e-32
TC81568 similar to GP|13277342|emb|CAC34417. Germin-like protein... 132 1e-31
TC82327 weakly similar to SP|O04012|ABPB_PRUPE Auxin-binding pro... 129 8e-31
BQ147886 weakly similar to GP|7242813|emb germin-like protein {P... 125 1e-29
CA989865 weakly similar to GP|7242813|emb germin-like protein {P... 125 2e-29
>TC76909 similar to SP|Q9S8P4|RHRE_PEA Rhicadhesin receptor precursor
(Germin-like protein). [Garden pea] {Pisum sativum},
partial (92%)
Length = 1054
Score = 416 bits (1070), Expect = e-117
Identities = 213/213 (100%), Positives = 213/213 (100%)
Frame = +2
Query: 1 MKIIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAK 60
MKIIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAK
Sbjct: 101 MKIIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAK 280
Query: 61 PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQL 120
PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQL
Sbjct: 281 PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQL 460
Query: 121 DVGFITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTL 180
DVGFITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTL
Sbjct: 461 DVGFITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTL 640
Query: 181 FASTPPVPDNILTQAFQIGTKEVQKIKSRLAPK 213
FASTPPVPDNILTQAFQIGTKEVQKIKSRLAPK
Sbjct: 641 FASTPPVPDNILTQAFQIGTKEVQKIKSRLAPK 739
>TC88864 similar to GP|1934730|gb|AAB51752.1| germin-like protein
{Arabidopsis thaliana}, partial (91%)
Length = 926
Score = 319 bits (817), Expect = 6e-88
Identities = 157/206 (76%), Positives = 180/206 (87%)
Frame = +1
Query: 8 LLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNT 67
L L+L T+ ++DPD LQDLCVADLAS + NGFTCK AS V A DFS+ +LAKPG+TNNT
Sbjct: 100 LALLLATSFSSDPDYLQDLCVADLASGVTANGFTCKEASKVNAFDFSSIILAKPGSTNNT 279
Query: 68 FGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITT 127
FGS+VT ANVQK+PGLNTLGVSL+RIDYA G LNPPHTHPRATE+V+VLEGQLDVGFITT
Sbjct: 280 FGSVVTGANVQKVPGLNTLGVSLSRIDYAPGSLNPPHTHPRATEVVFVLEGQLDVGFITT 459
Query: 128 ANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPV 187
ANVLISKTI KGE FVFPKGLVHFQKN+ VPA+V++ FNSQL GT +I TLFA+TP V
Sbjct: 460 ANVLISKTISKGEIFVFPKGLVHFQKNNANVPASVLSAFNSQLPGTQSIATTLFAATPSV 639
Query: 188 PDNILTQAFQIGTKEVQKIKSRLAPK 213
PDN+LT+ FQ+GTKEV+KIKSRLAPK
Sbjct: 640 PDNVLTKTFQVGTKEVEKIKSRLAPK 717
>TC79592 similar to GP|1934730|gb|AAB51752.1| germin-like protein
{Arabidopsis thaliana}, partial (96%)
Length = 947
Score = 275 bits (702), Expect = 1e-74
Identities = 138/210 (65%), Positives = 166/210 (78%)
Frame = +3
Query: 4 IAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGA 63
I V+L ++ T+ +DPD LQDLCVA +S + VNGF CK SNVTA DF + LA P
Sbjct: 117 ILVILCCAISFTIASDPDTLQDLCVALPSSGVKVNGFACKEESNVTAVDFFFDGLANPKV 296
Query: 64 TNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVG 123
NNT GSLVT ANV KIPGLNTLGVSL+RIDY GLNPPHTHPRATE+V+VLEG+LDVG
Sbjct: 297 VNNTVGSLVTAANVDKIPGLNTLGVSLSRIDYKPKGLNPPHTHPRATEVVFVLEGELDVG 476
Query: 124 FITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFAS 183
FITT+N LISK+I KGE FVFPKGLVH+QKN+G A+VI+ FNSQL GT++I +LF S
Sbjct: 477 FITTSNKLISKSIKKGEIFVFPKGLVHYQKNNGDKAASVISAFNSQLPGTLSIVSSLFDS 656
Query: 184 TPPVPDNILTQAFQIGTKEVQKIKSRLAPK 213
TP VPD++L QAFQI K+V +IK+++APK
Sbjct: 657 TPTVPDDVLAQAFQIDAKQVDEIKTKVAPK 746
>AW691378 similar to SP|Q9S8P4|RHRE Rhicadhesin receptor precursor
(Germin-like protein). [Garden pea] {Pisum sativum},
partial (63%)
Length = 659
Score = 236 bits (601), Expect = 6e-63
Identities = 136/192 (70%), Positives = 147/192 (75%), Gaps = 5/192 (2%)
Frame = +1
Query: 1 MKIIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAK 60
MKIIAVLLLLVLTTTVTADPDNLQDLCVADLAS VNGFTCKPASNVTAADFSTNVLAK
Sbjct: 91 MKIIAVLLLLVLTTTVTADPDNLQDLCVADLAS---VNGFTCKPASNVTAADFSTNVLAK 261
Query: 61 PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQL 120
PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEI +
Sbjct: 262 PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEICLCS*RSI 441
Query: 121 DVGFITTANVL-ISKTIVKGETFVFPKGLVHFQKNSG--YVPAAVIAGFNSQLQGTVNIP 177
+ + + ++KGETFVFPK + F K + YVPAAVIA + I
Sbjct: 442 RCWVHNNS*MF*YQRLLLKGETFVFPKRIGSFSKRNSWVYVPAAVIARIQ*SIAKEQ*IF 621
Query: 178 LT--LFASTPPV 187
L ++ASTPPV
Sbjct: 622 LLNFVYASTPPV 657
>TC78488 similar to GP|6689036|emb|CAB65370.1 germin-like protein {Pisum
sativum}, partial (98%)
Length = 882
Score = 177 bits (448), Expect = 3e-45
Identities = 103/212 (48%), Positives = 128/212 (59%), Gaps = 5/212 (2%)
Frame = +2
Query: 3 IIAVLLLLVLTTTVTA-DPDNLQDLCVA--DLASAILVNGFTCKPASNVTAADFSTNVLA 59
I+ V L L + A DP LQD CVA D + + VNG CK T DF +V
Sbjct: 44 ILFVTAFLALASCAFAFDPSPLQDFCVAINDTKNGVFVNGKFCKDPKLATPNDFFFSV-- 217
Query: 60 KPGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQ 119
K G +N GS VT V I GLNTLG+SLARID+A GLNPPHTHPRATEI+ VLEG
Sbjct: 218 KEGNISNPLGSKVTPVTVNDILGLNTLGISLARIDFASRGLNPPHTHPRATEILIVLEGT 397
Query: 120 LDVGFITT--ANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIP 177
L VGF+T+ N LI+K + KG+ FVFP GL+HFQ N GY A IAG +SQ G + I
Sbjct: 398 LYVGFVTSNPENRLITKVLNKGDVFVFPIGLIHFQLNVGYGNAVAIAGLSSQNPGVITIA 577
Query: 178 LTLFASTPPVPDNILTQAFQIGTKEVQKIKSR 209
+F S P + +LT+AFQ+ V ++ +
Sbjct: 578 NAVFGSNPKISSEVLTKAFQVDNNIVDNLQKQ 673
>TC85455 similar to GP|6689036|emb|CAB65370.1 germin-like protein {Pisum
sativum}, partial (97%)
Length = 929
Score = 176 bits (446), Expect = 6e-45
Identities = 99/211 (46%), Positives = 126/211 (58%), Gaps = 5/211 (2%)
Frame = +1
Query: 4 IAVLLLLVLTTTVTADPDNLQDLCVA--DLASAILVNGFTCKPASNVTAADFSTNVLAKP 61
I LL L + DP LQD CVA D + + VNG CK A DF L P
Sbjct: 70 IVALLALASSVGFAYDPSPLQDFCVAINDTKTGVFVNGKFCKDPKLANADDFFFQGLG-P 246
Query: 62 GATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLD 121
G T+N GS VT V +I GLNTLG+SLAR+D+A GLNPPHTHPR TEI+ VLEG L
Sbjct: 247 GNTSNPLGSKVTPVTVNEILGLNTLGISLARVDFAPKGLNPPHTHPRGTEILVVLEGTLY 426
Query: 122 VGFITT---ANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPL 178
VGF+++ N L +K + KG+ FVFP GL+HFQ N GY A IAG +SQ G + I
Sbjct: 427 VGFVSSNQDNNRLFTKVLNKGDVFVFPIGLIHFQLNVGYGEAIAIAGLSSQNPGVITIAN 606
Query: 179 TLFASTPPVPDNILTQAFQIGTKEVQKIKSR 209
+F S PP+ +LT+AFQ+ + ++ +
Sbjct: 607 AVFGSKPPISLEVLTKAFQVDKNVIDYLQKQ 699
>AW688621 weakly similar to SP|P92998|GL11 Germin-like protein subfamily 1
member 1 precursor. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (65%)
Length = 644
Score = 172 bits (437), Expect = 7e-44
Identities = 88/190 (46%), Positives = 118/190 (61%), Gaps = 1/190 (0%)
Frame = +2
Query: 18 ADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLVTLANV 77
+DPD LQD C+AD + +NG C T++ F+T+ L+KPG T N FG VT N
Sbjct: 74 SDPDPLQDYCIADNKNTFFLNGLPCIDPKQATSSHFATSSLSKPGNTTNMFGFSVTPTNT 253
Query: 78 QKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITTANVLISKTIV 137
+PGLNTLG+ L R+D A G+ PPH+HPRA+E+ ++G L VGF+ T+N ++ +
Sbjct: 254 INLPGLNTLGLVLVRVDIAGNGIVPPHSHPRASEVTTCIKGLLLVGFVDTSNRAFTQNLS 433
Query: 138 KGETFVFPKGLVHFQKN-SGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNILTQAF 196
GE+FVFPKGLVHF N PA I+G NSQ G + FAS P +PD +L +AF
Sbjct: 434 PGESFVFPKGLVHFLYNRDSKQPAIAISGLNSQNPGAQIASIATFASKPSIPDEVLKKAF 613
Query: 197 QIGTKEVQKI 206
QI +EV I
Sbjct: 614 QINGQEVDII 643
>TC87484 homologue to GP|6689036|emb|CAB65370.1 germin-like protein {Pisum
sativum}, complete
Length = 950
Score = 170 bits (431), Expect = 3e-43
Identities = 99/206 (48%), Positives = 126/206 (61%), Gaps = 8/206 (3%)
Frame = +2
Query: 1 MKIIAVLLLLVLTTTVTA---DPDNLQDLCVA--DLASAILVNGFTCKPASNVTAADFST 55
MKI+ L+ ++ + A DP LQD CVA D + VNG CK + V A DF
Sbjct: 71 MKILYFLVSILALASSLAFAYDPSPLQDFCVAIKDPKDGVFVNGKFCKDPALVKAEDFFK 250
Query: 56 NVLAKPGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYV 115
+V A G +N GS VT V ++ GLNTLG+SLARID+ GLNPPH HPR TEI+ V
Sbjct: 251 HVEA--GNASNALGSQVTPVTVDQLFGLNTLGISLARIDFVPRGLNPPHIHPRGTEILIV 424
Query: 116 LEGQLDVGFITT---ANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQG 172
LEG L VGF+T+ N L +K + KG+ FVFP GL+HFQ N GY A IAG +SQ G
Sbjct: 425 LEGTLYVGFVTSNQDNNRLFTKVLNKGDVFVFPIGLIHFQLNVGYGNAVAIAGLSSQNPG 604
Query: 173 TVNIPLTLFASTPPVPDNILTQAFQI 198
+ + LF S P + D +LT+AFQ+
Sbjct: 605 VITVANALFKSNPLISDEVLTKAFQV 682
>TC87485 homologue to GP|6689036|emb|CAB65370.1 germin-like protein {Pisum
sativum}, complete
Length = 888
Score = 169 bits (428), Expect = 7e-43
Identities = 94/185 (50%), Positives = 117/185 (62%), Gaps = 5/185 (2%)
Frame = +2
Query: 19 DPDNLQDLCVA--DLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLVTLAN 76
DP LQD CVA D + VNG CK + V A DF +V A G +N GS VT
Sbjct: 17 DPSPLQDFCVAIKDPKDGVFVNGKFCKDPALVKAEDFFKHVEA--GNASNALGSQVTPVT 190
Query: 77 VQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITT---ANVLIS 133
V ++ GLNTLG+SLAR+D+A GLNPPH HPR TEI+ VLEG L VGF+T+ N L +
Sbjct: 191 VDQLFGLNTLGISLARVDFAPKGLNPPHIHPRGTEILIVLEGTLYVGFVTSNQDNNRLFT 370
Query: 134 KTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNILT 193
K + KG+ FVFP GL+HFQ N GY A IAG +SQ G + + LF S P + D +LT
Sbjct: 371 KVLNKGDVFVFPIGLIHFQLNVGYGNAVAIAGLSSQNPGVITVANALFKSDPLISDEVLT 550
Query: 194 QAFQI 198
+AFQ+
Sbjct: 551 KAFQV 565
>AJ498555 weakly similar to SP|Q9FMA8|GL1B Germin-like protein subfamily 1
member 11 precursor. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (65%)
Length = 499
Score = 146 bits (369), Expect = 5e-36
Identities = 83/159 (52%), Positives = 107/159 (67%), Gaps = 5/159 (3%)
Frame = +2
Query: 19 DPDNLQDLCVA--DLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLVTLAN 76
D LQD CVA D+ + +LVNG CK VTA DF +V K G ++ G+ VTL
Sbjct: 26 DLSPLQDFCVATNDIKTGVLVNGQYCKDPKLVTADDFFFSV--KEGVVSSPVGTQVTLVT 199
Query: 77 VQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITT---ANVLIS 133
V +I GLNTLGVSLARID+A G+NPPHTHPRATEI+ VL+G L+VGF T+ ++ I+
Sbjct: 200 VNEILGLNTLGVSLARIDFAPKGINPPHTHPRATEILMVLDGTLNVGFYTSNQESSTPIT 379
Query: 134 KTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQG 172
K + KG+ VFP GL+HF+ N+G A I+GF+SQ G
Sbjct: 380 KDLNKGDVSVFPIGLIHFEHNTGDGNAVAISGFSSQNPG 496
>TC81825 weakly similar to SP|Q9FIC8|GL1G_ARATH Germin-like protein
subfamily 1 member 16 precursor. [Mouse-ear cress]
{Arabidopsis thaliana}, partial (76%)
Length = 1042
Score = 146 bits (368), Expect = 7e-36
Identities = 78/176 (44%), Positives = 107/176 (60%), Gaps = 2/176 (1%)
Frame = +3
Query: 36 LVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDY 95
L+ CK V A DF + L G T N GS VT ++PGLNTLG+S+AR+D
Sbjct: 234 LLGNSLCKDPKLVEANDFFFSGLHIAGNTTNAAGSKVTPVFAAQLPGLNTLGISMARVDI 413
Query: 96 AVGGLNPPHTHPRATEIVYVLEGQLDVGFITT--ANVLISKTIVKGETFVFPKGLVHFQK 153
A G+NPPH+HPRATEI VLEG L+VGFIT+ N SK + KG+ FVFP GL+H+Q+
Sbjct: 414 APWGVNPPHSHPRATEIFTVLEGTLEVGFITSNPENRHFSKVLHKGDVFVFPIGLIHYQR 593
Query: 154 NSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNILTQAFQIGTKEVQKIKSR 209
N G+ IA +SQ G + I +F +TP + +L +AFQ+ + ++S+
Sbjct: 594 NIGHDNVIAIAALSSQNPGAITIGNAVFGATPEIASEVLIKAFQLEKNAINYLQSK 761
>TC76324 similar to GP|7242813|emb|CAB77393.1 germin-like protein {Phaseolus
vulgaris}, partial (93%)
Length = 886
Score = 143 bits (361), Expect = 4e-35
Identities = 81/209 (38%), Positives = 124/209 (58%)
Frame = +1
Query: 1 MKIIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAK 60
MKII ++ L + T+ ++ D CVADL + +G+ CKP +N+T+ DF + K
Sbjct: 55 MKIIHIVFLFSFLSFTTSQA-SVNDFCVADLKAPDTPSGYHCKPLANITSDDFVFHGF-K 228
Query: 61 PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQL 120
G TNN+F + +T A V PGLN LG+S AR+D A G P HTHP ATE++ +++G++
Sbjct: 229 AGNTNNSFNAALTSAFVTDFPGLNGLGISAARLDIAENGSIPMHTHPGATELLIIVQGEI 408
Query: 121 DVGFITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTL 180
GF+T V SKT+ G+ VFP+G++HFQ N+G A F+S G + L L
Sbjct: 409 TAGFLTPTAV-YSKTLKPGDLMVFPQGMLHFQINTGKGKATAFLTFSSANPGAQLLDLLL 585
Query: 181 FASTPPVPDNILTQAFQIGTKEVQKIKSR 209
F++ +P ++ Q + +VQK+K+R
Sbjct: 586 FSNN--LPSQLVAQTTFLDLAQVQKLKAR 666
>TC79767 similar to SP|Q9SR72|GL32_ARATH Germin-like protein subfamily 3
member 2 precursor. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (84%)
Length = 965
Score = 142 bits (359), Expect = 7e-35
Identities = 79/203 (38%), Positives = 118/203 (57%), Gaps = 7/203 (3%)
Frame = +3
Query: 18 ADPDNLQDLCVAD--LASAILVNG-----FTCKPASNVTAADFSTNVLAKPGATNNTFGS 70
+DPD +QD C+ + LAS I + CK +S V DF + + G + T G
Sbjct: 21 SDPDPVQDFCIPNPILASMIKTHHTFHTILPCKNSSEVITNDFIFSNMKTSGNFSET-GL 197
Query: 71 LVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITTANV 130
V AN PGLNTLG+S AR D +GG+NPPH HPRATE+++V++G++ GF+ + N
Sbjct: 198 AVMPANPTNFPGLNTLGMSFARTDIEIGGINPPHFHPRATELIHVIQGKVYSGFVDSNNK 377
Query: 131 LISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDN 190
+ ++ + +GE VFP+GLVHF N G + FNSQ G IP +F S + +
Sbjct: 378 VFARILEQGEVMVFPRGLVHFMMNVGDEVVTLFGSFNSQNPGLQKIPSAVFGS--GIDEE 551
Query: 191 ILTQAFQIGTKEVQKIKSRLAPK 213
+L +AF + +K++ +K +L PK
Sbjct: 552 LLQKAFGLSSKQIGTMKRKLDPK 620
>TC86278 similar to GP|17473703|gb|AAL38307.1 germin-like protein
{Arabidopsis thaliana}, partial (88%)
Length = 908
Score = 141 bits (356), Expect = 2e-34
Identities = 77/205 (37%), Positives = 119/205 (57%)
Frame = +2
Query: 6 VLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATN 65
+ ++ +L+ ++ ++QD CVAD+ + +G+ CKPAS VT+ DF+ L PG
Sbjct: 47 IQIIFLLSLFLSISNASVQDFCVADIKGSDTPSGYPCKPASTVTSDDFAFEGLIAPGNIT 226
Query: 66 NTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFI 125
N + VT A V + P +N LG+S AR+D G+ P HTHP A+E++ V +G + GF+
Sbjct: 227 NIINAAVTPAFVAQFPAVNGLGLSAARLDLGPAGVIPLHTHPGASELLVVTQGHITAGFV 406
Query: 126 TTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTP 185
++AN + KT+ KGE VFP+GL+HFQ +G A A F+S G + LFAS
Sbjct: 407 SSANTVYIKTLKKGELMVFPQGLLHFQATAGKRNAVAFAVFSSASPGLQILDFALFASNF 586
Query: 186 PVPDNILTQAFQIGTKEVQKIKSRL 210
P ++T+ + V+K+KS L
Sbjct: 587 STP--LITKTTFLDPVLVKKLKSIL 655
>TC76383 similar to GP|7242813|emb|CAB77393.1 germin-like protein {Phaseolus
vulgaris}, partial (93%)
Length = 956
Score = 139 bits (351), Expect = 6e-34
Identities = 79/209 (37%), Positives = 121/209 (57%)
Frame = +3
Query: 1 MKIIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAK 60
MKI +L L L ++ T+ ++ D CV D+ + G+ C P +N+T+ DF +
Sbjct: 96 MKITPILFLFTLLSSTTSYA-SVHDFCVGDIKAPETPTGYHCMPLANITSDDFVFHGFVA 272
Query: 61 PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQL 120
G TNN+F + +T A V P LN LG+S AR+D A GG P HTHP ATE++ ++ G++
Sbjct: 273 -GNTNNSFNAALTSAFVTDFPALNDLGISAARLDIAKGGSIPMHTHPGATELLIMVHGEI 449
Query: 121 DVGFITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTL 180
GF+TT V SKT+ G+ VFP+G++HFQ NSG A F+S G + L L
Sbjct: 450 TAGFLTTTAV-YSKTLKPGDLMVFPQGMLHFQVNSGKGKATAFLTFSSANPGAQLLDLLL 626
Query: 181 FASTPPVPDNILTQAFQIGTKEVQKIKSR 209
F++ +P ++ Q + +V+K+K+R
Sbjct: 627 FSNN--LPSELVAQTTFLDLAQVKKLKAR 707
>TC85909 similar to GP|13277342|emb|CAC34417. Germin-like protein {Pisum
sativum}, partial (95%)
Length = 1551
Score = 133 bits (334), Expect = 6e-32
Identities = 80/208 (38%), Positives = 116/208 (55%)
Frame = +2
Query: 3 IIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPG 62
I+A+ + L + A ++ D CVAD G++CK S VT DF+ + LA G
Sbjct: 29 IVAISFIFSLLSLSHA---SVVDFCVADYNLPNGPAGYSCKTPSKVTENDFAYHGLAAAG 199
Query: 63 ATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDV 122
T+N + VT A + GLN LG+SLAR+D A GG+ P HTHP A+E++ V++G +
Sbjct: 200 NTSNIIKAAVTPAFDAQFAGLNGLGISLARLDLAAGGVIPLHTHPGASEVLVVIQGTICA 379
Query: 123 GFITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFA 182
GF+++AN + KT+ KG+ VFP+GL+HFQ NSG A F+S G + LF
Sbjct: 380 GFVSSANTVYIKTLYKGDVMVFPQGLLHFQINSGGSNALAFVSFSSANPGLQILDFALFK 559
Query: 183 STPPVPDNILTQAFQIGTKEVQKIKSRL 210
S P ++T I V+K+K L
Sbjct: 560 S--DFPTELITATTFIDAAVVKKLKGVL 637
>TC81568 similar to GP|13277342|emb|CAC34417. Germin-like protein {Pisum
sativum}, partial (96%)
Length = 684
Score = 132 bits (331), Expect = 1e-31
Identities = 75/205 (36%), Positives = 115/205 (55%)
Frame = +3
Query: 6 VLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATN 65
+L + + + ++ ++ D CVAD + G++CKP S VT DF + LA G T
Sbjct: 12 ILTIFFIFSLLSLAHASVVDFCVADYNAPNGPAGYSCKPPSKVTVNDFVYHGLAAAGNTT 191
Query: 66 NTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFI 125
N + VT A + PG+N LG+S+AR+D AVGG+ P HTHP A+E++ V++G + GF+
Sbjct: 192 NIIKAAVTTAIDAQFPGVNGLGISIARLDIAVGGVIPLHTHPGASEVLVVIKGTISAGFV 371
Query: 126 TTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTP 185
++ NV+ KT+ KG+ +FP+GL+HFQ N G A F+S + LF S
Sbjct: 372 SSDNVVYLKTLHKGDVMIFPQGLLHFQINVGGSNALTFNSFSSANPRLQILDYALFES-- 545
Query: 186 PVPDNILTQAFQIGTKEVQKIKSRL 210
P ++T I V+K+K L
Sbjct: 546 DFPTKLITATTFIDPAVVKKLKGIL 620
>TC82327 weakly similar to SP|O04012|ABPB_PRUPE Auxin-binding protein ABP19b
precursor. [Peach] {Prunus persica}, partial (73%)
Length = 734
Score = 129 bits (324), Expect = 8e-31
Identities = 80/199 (40%), Positives = 112/199 (56%)
Frame = +1
Query: 11 VLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGS 70
VL ++ T+ D DLCVADL +G+ CK +NVT DF + L PG+T N F
Sbjct: 1 VLLSSYTSYAD---DLCVADLLLPNTPSGYPCKSETNVTVNDFVFSGLV-PGSTINPFNF 168
Query: 71 LVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITTANV 130
+T A V +PGLN LG+S AR D+ + G P HTH ATE++ V+EGQ+ GFIT V
Sbjct: 169 AITSAFVTSLPGLNGLGISAARADFGINGSVPVHTHD-ATELLIVVEGQITAGFITRTKV 345
Query: 131 LISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDN 190
SKTI G+ VFPKGL+HF NSG A F+S T + LF + + +
Sbjct: 346 -YSKTIKPGDLIVFPKGLLHFVVNSGVGKAVAFVAFSSSNPSTQILDTLLFGNN--LSTS 516
Query: 191 ILTQAFQIGTKEVQKIKSR 209
I+ + + ++ K+K++
Sbjct: 517 IIAETTLLDVSQILKLKAQ 573
>BQ147886 weakly similar to GP|7242813|emb germin-like protein {Phaseolus
vulgaris}, partial (73%)
Length = 646
Score = 125 bits (314), Expect = 1e-29
Identities = 70/188 (37%), Positives = 107/188 (56%)
Frame = +1
Query: 22 NLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLVTLANVQKIP 81
++ D CVADL + +G+ CKP ++VT+ DFS + L TNN+F V A V P
Sbjct: 64 SINDFCVADLKAPNTPSGYACKPLASVTSDDFSFHGLVAAN-TNNSFKIGVATATVTNFP 240
Query: 82 GLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITTANVLISKTIVKGET 141
LN LG+S RID GL P HTHP ATE++ V++G++ GF+T + SK + G+
Sbjct: 241 ALNGLGISALRIDLDQDGLAPMHTHPDATELLSVVKGEITAGFLTPTS-FYSKVLKSGDV 417
Query: 142 FVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNILTQAFQIGTK 201
FVFP+G++HF NSG A F+S+ T + + LF + +P + +Q +
Sbjct: 418 FVFPQGMLHFAVNSGKGKATAFGAFSSENPTTHILDVLLFGN--KLPSGLXSQTTLLDLA 591
Query: 202 EVQKIKSR 209
+V +K++
Sbjct: 592 QVXXLKAK 615
>CA989865 weakly similar to GP|7242813|emb germin-like protein {Phaseolus
vulgaris}, partial (93%)
Length = 729
Score = 125 bits (313), Expect = 2e-29
Identities = 70/188 (37%), Positives = 109/188 (57%)
Frame = +2
Query: 22 NLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLVTLANVQKIP 81
++ D CVADL + +G+ CKP +++T+ DF + L G T+N+F VT A V P
Sbjct: 23 SVNDFCVADLKAPNTNSGYPCKPVASITSDDFVFHGLVA-GNTSNSFKIGVTSATVTNFP 199
Query: 82 GLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITTANVLISKTIVKGET 141
LN LG+S R+D GGL+P HTHP ATE+ V++G+ GF+T + SK + G+
Sbjct: 200 ALNGLGISAVRVDMEEGGLSPMHTHPDATELGIVVQGEFTAGFLTPTS-FYSKVLKAGDV 376
Query: 142 FVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNILTQAFQIGTK 201
FV PKG++HF NSG A F+S+ + LFA+ +P +++ + +
Sbjct: 377 FVVPKGMLHFAINSGKGKAIGYVSFSSENPTIHTLDSLLFAN--KLPSDLVAKTTLLDID 550
Query: 202 EVQKIKSR 209
+V+K+K+R
Sbjct: 551 QVKKLKAR 574
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,580,449
Number of Sequences: 36976
Number of extensions: 64602
Number of successful extensions: 372
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 334
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 334
length of query: 213
length of database: 9,014,727
effective HSP length: 92
effective length of query: 121
effective length of database: 5,612,935
effective search space: 679165135
effective search space used: 679165135
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149038.15


