

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.5 + phase: 0
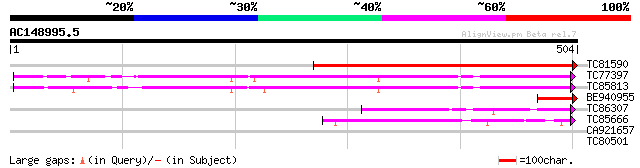
(504 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81590 similar to GP|6041790|gb|AAF02110.1| putative NADPH-ferr... 448 e-126
TC77397 homologue to PIR|S37159|S37159 NADPH--ferrihemoprotein r... 233 2e-61
TC85813 homologue to GP|6503253|gb|AAC09468.2| putative NADPH-cy... 219 3e-57
BE940955 74 2e-13
TC86307 homologue to PIR|T06773|T06773 ferredoxin--NADP+ reducta... 62 4e-10
TC85666 homologue to SP|P10933|FENR_PEA Ferredoxin--NADP reducta... 62 5e-10
CA921657 similar to PIR|T03826|T03 cytochrome b245 beta chain ho... 28 6.3
TC80501 similar to PIR|T52458|T52458 chlorophyll b synthase [imp... 28 8.3
>TC81590 similar to GP|6041790|gb|AAF02110.1| putative
NADPH-ferrihemoprotein reductase {Arabidopsis thaliana},
partial (33%)
Length = 1095
Score = 448 bits (1153), Expect = e-126
Identities = 213/234 (91%), Positives = 223/234 (95%)
Frame = +3
Query: 271 LIQLVPMLKKREFSISSSQSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAV 330
LIQLVPMLK R FSISSSQS HPNQVHLTVSVVSWTTPYKRKKKGLCSSWLA LDPR+AV
Sbjct: 3 LIQLVPMLKTRAFSISSSQSVHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLATLDPRNAV 182
Query: 331 SLPVWFQKGSLPTPSPSLPLILVGPGTGCAPFRGFIEERALQSKTISIAPIIFFFGCWNE 390
+P WFQKGSLPT SPSLPLILVGPGTGCAPFRGFIEERALQSKTIS API+FFFGCWNE
Sbjct: 183 GIPAWFQKGSLPTASPSLPLILVGPGTGCAPFRGFIEERALQSKTISTAPIMFFFGCWNE 362
Query: 391 DGDFLYKDFWLNHSQNNGVLSESTGGGFYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEG 450
DGDFLYKD WLNHSQNNGVLSE+ GGGFYVAFSRDQPEKVYVQHK++EHSGRVWNLLA+G
Sbjct: 363 DGDFLYKDLWLNHSQNNGVLSEANGGGFYVAFSRDQPEKVYVQHKMKEHSGRVWNLLADG 542
Query: 451 ASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAWS 504
A+VYIAGS TKMPTDVTSAFEEIVSKEN+VSKEDAVRWIRALE+CGKYHIEAWS
Sbjct: 543 AAVYIAGSSTKMPTDVTSAFEEIVSKENEVSKEDAVRWIRALERCGKYHIEAWS 704
>TC77397 homologue to PIR|S37159|S37159 NADPH--ferrihemoprotein reductase (EC
1.6.2.4) - spring vetch, complete
Length = 2519
Score = 233 bits (593), Expect = 2e-61
Identities = 166/527 (31%), Positives = 256/527 (48%), Gaps = 27/527 (5%)
Frame = +2
Query: 4 VTKKLDKRLMDLGGKAILERGLGDDQQPSGYEGTLDPWLSSLWRMLNMIKPELLPSGPDV 63
+ K +D L + G K ++ G+GDD Q E + W SLW L+ +LL DV
Sbjct: 662 IGKVVDDDLSEQGAKRLVPLGMGDDDQ--SIEDDFNAWKESLWPELD----QLLRDEDDV 823
Query: 64 LIQDT----------TLIDQPKVQITYHNIENVVSHSSDTGSVRSMHPGKSSSNRNGYPD 113
T + P V +Y N N +++ G+V +H +P
Sbjct: 824 NTVSTPYTAAISEYRVVFHDPTVTPSYENHFN----AANGGAVFDIH----------HP- 958
Query: 114 CFLKLVKNLPLTKPNSGKDVRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNL 173
C + L KP S + H EF+ + Y+TGD + V V +
Sbjct: 959 CRANVAVRRELHKPQSDRSCIHLEFDVSGTGVTYETGDHVGVYADNCDETVKEAGKLLGQ 1138
Query: 174 DPDSLITVSPKGMDCNGHGSRM------PVKLRTFVELTMDVASASPRR---YFFEARCS 224
D D L ++ D G + P +RT + D+ + PR+ A S
Sbjct: 1139 DLDLLFSLHTDNEDGTSLGGSLLPPFPGPCTVRTALARYADLLNP-PRKAALIALAAHAS 1315
Query: 225 E-HERERLEYFASPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPLE-WLIQLVPMLKKRE 282
E E ERL++ +SP+G+D+ ++ RT+LEV+ DFPS + PL + + P L+ R
Sbjct: 1316 EPSEAERLKFLSSPQGKDEYSKWVVGSHRTLLEVMADFPSAKPPLGVFFAAIAPRLQPRY 1495
Query: 283 FSISSSQSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDP----RDAVSLPVWFQK 338
+SISSS P +VH+T ++V TP R KG+CS+W+ P RD P++ +
Sbjct: 1496 YSISSSPRFAPQRVHVTCALVEGPTPTGRIHKGVCSTWMKNAIPSEESRDCSWAPIFIRP 1675
Query: 339 GSLPTPS-PSLPLILVGPGTGCAPFRGFIEER-ALQSKTISIAPIIFFFGCWNEDGDFLY 396
+ P+ PS+P+I+VGPGTG APFRGF++ER AL+ + + P + FFGC N DF+Y
Sbjct: 1676 SNFKLPADPSIPIIMVGPGTGLAPFRGFLQERFALKEDGVQLGPALLFFGCRNRQMDFIY 1855
Query: 397 KDFWLNHSQNNGVLSESTGGGFYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEGASVYIA 456
++ LN+ G LSE VAFSR+ PEK YVQHK+ + + W+L+++G +Y+
Sbjct: 1856 EEE-LNNFVEQGSLSE-----LIVAFSREGPEKEYVQHKMMDKASYFWSLISQGGYLYVC 2017
Query: 457 GSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAW 503
G M DV IV ++ + A ++ L+ G+Y + W
Sbjct: 2018 GDAKGMARDVHRTLHTIVQQQENADSSKAEATVKKLQMDGRYLRDVW 2158
>TC85813 homologue to GP|6503253|gb|AAC09468.2| putative NADPH-cytochrome P450
reductase {Pisum sativum}, complete
Length = 2581
Score = 219 bits (557), Expect = 3e-57
Identities = 154/523 (29%), Positives = 251/523 (47%), Gaps = 23/523 (4%)
Frame = +3
Query: 4 VTKKLDKRLMDLGGKAILERGLGDDQQPSGYEGTLDPWLSSLWRMLNMIKPE------LL 57
V K +D +L++ GG ++ GLGDD Q E W LW L+ + +
Sbjct: 711 VAKIVDDKLLEKGGNRLVPVGLGDDDQC--IEDDFTAWKEELWPALDQLLRDEDDATVAT 884
Query: 58 PSGPDVLIQDTTLIDQPKVQITYHNIENVVSHSSDTGSVRSMHPGKSSSNRNGYPDCFLK 117
P VL + DQ + N H+ V + HP +++
Sbjct: 885 PYTASVLEYRVVIRDQLDATVDEKKQLNGNGHAV----VDAHHPVRAN------------ 1016
Query: 118 LVKNLPLTKPNSGKDVRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDS 177
+ L P S + H EF+ + Y+TGD + V S V+ R L PD+
Sbjct: 1017 VAVRKELHTPASDRSCTHLEFDISGTGVVYETGDHVGVYCENLSDTVEEAERILGLSPDT 1196
Query: 178 LITVSPKGMDCNGHGSRM------PVKLRTFVELTMDVASASPRRYFFEARCSE----HE 227
+V D G P LRT + DV S SP++ A + E
Sbjct: 1197 YFSVHTDDEDGKPLGGSSLPPPFPPCTLRTALAKYADVLS-SPKKSALLALAAHASDPSE 1373
Query: 228 RERLEYFASPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPLE-WLIQLVPMLKKREFSIS 286
+RL + ASP G+D+ ++ +R++LEV+ +F S + P+ + + P L+ R +SIS
Sbjct: 1374 ADRLRHLASPAGKDEYAEWVIASQRSLLEVMAEFSSAKPPIGVFFASVAPRLQPRYYSIS 1553
Query: 287 SSQSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDP----RDAVSLPVWFQKGSLP 342
SS P+++H+T ++V P R +G+CS+W+ P +D P++ ++ +
Sbjct: 1554 SSPRVAPSRIHVTCALVHDKMPTGRIHQGVCSTWMKNSAPLEKSQDCSWAPIFVRQSNFR 1733
Query: 343 TPSPS-LPLILVGPGTGCAPFRGFIEER-ALQSKTISIAPIIFFFGCWNEDGDFLYKDFW 400
P+ + +P+I++GPGTG APFRGF++ER AL+ + + P + FFGC N D++Y+D
Sbjct: 1734 LPADNKVPIIMIGPGTGLAPFRGFLQERLALKEEGAELGPSVLFFGCRNRQVDYIYEDE- 1910
Query: 401 LNHSQNNGVLSESTGGGFYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLT 460
LNH + G LSE VAFSR+ P K YVQHK+ E + +WN++++GA +Y+ G
Sbjct: 1911 LNHFVHGGALSE-----LIVAFSREGPTKEYVQHKMIEKASDIWNMISQGAYIYVCGDAK 2075
Query: 461 KMPTDVTSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAW 503
M DV I+ ++ + ++ L+ G+Y + W
Sbjct: 2076 GMAKDVHRTLHTILQEQGSLDNSKTESMVKNLQMTGRYLRDVW 2204
>BE940955
Length = 337
Score = 73.6 bits (179), Expect = 2e-13
Identities = 34/35 (97%), Positives = 34/35 (97%)
Frame = +2
Query: 470 FEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAWS 504
FEEIVSKENDVSKE AVRWIRALEKCGKYHIEAWS
Sbjct: 2 FEEIVSKENDVSKEYAVRWIRALEKCGKYHIEAWS 106
>TC86307 homologue to PIR|T06773|T06773 ferredoxin--NADP+ reductase (EC
1.18.1.2) - garden pea (fragment), complete
Length = 1648
Score = 62.4 bits (150), Expect = 4e-10
Identities = 52/198 (26%), Positives = 90/198 (45%), Gaps = 7/198 (3%)
Frame = +3
Query: 313 KKGLCSSWLAALDPRDAVSLPVWFQKGSL-PTPSPSLPLILVGPGTGCAPFRGFIEERAL 371
K G+CS++L P D + + K L P P+ I++ GTG AP+RG++ +
Sbjct: 822 KNGVCSNFLCDSKPGDKIKITGPSGKIMLLPEDDPNATHIMIATGTGVAPYRGYLRRMFM 1001
Query: 372 QS-KTISIAPIIF-FFGCWNEDGDFLYKDFWLNHSQNNGVLSESTGGGFYVAFSRDQPE- 428
+S T + + F G N D LY D + + ++ + A SR++
Sbjct: 1002 ESVPTFKFGGLAWLFLGVANSD-SLLYDDEFTKYLKD-----YPENFRYDRALSREEKNR 1163
Query: 429 ---KVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDA 485
K+YVQ K+ E+S ++ LL GA +Y G MP +E + + + E
Sbjct: 1164 NGGKMYVQDKIEEYSDEIFKLLDNGAHIYFCGLKGMMP-----GIQETLKRVAENRGESW 1328
Query: 486 VRWIRALEKCGKYHIEAW 503
+ L+K ++H+E +
Sbjct: 1329 EEKLSQLKKNKQWHVEVY 1382
>TC85666 homologue to SP|P10933|FENR_PEA Ferredoxin--NADP reductase leaf
isozyme chloroplast precursor (EC 1.18.1.2) (FNR).
[Garden pea], partial (96%)
Length = 1389
Score = 62.0 bits (149), Expect = 5e-10
Identities = 59/239 (24%), Positives = 104/239 (42%), Gaps = 14/239 (5%)
Frame = +3
Query: 279 KKREFSISSS---QSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVW 335
K R +SI+SS V L V + +T KG+CS++L L P V +
Sbjct: 501 KLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDAGEVVKGVCSNFLCDLRPGSEVQITGP 680
Query: 336 FQKGSLPTPSPSLPLILVGPGTGCAPFRGFIEERALQS-KTISIAPIIFFFGCWNEDGDF 394
K L P+ +I++G GTG APFR F+ + + + + + F
Sbjct: 681 VGKEMLMPKDPNATVIMLGTGTGIAPFRSFLWKMFFEKHEDYKFNGLAWLFLGVPTSSSL 860
Query: 395 LYKDFWLNHSQNNGVLSESTGGGFYVAFS------RDQPEKVYVQHKLREHSGRVWNLL- 447
LYK+ + + E F + F+ D+ EK+Y+Q ++ +++ +W LL
Sbjct: 861 LYKEEFEK-------MKEKAPENFRLDFAVSREQVNDKGEKMYIQTRMAQYAEELWELLK 1019
Query: 448 AEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWI---RALEKCGKYHIEAW 503
+ VY+ G L M + + +K D + WI R+L+K ++++E +
Sbjct: 1020KDNTFVYMCG-LKGMEKGIDDIMVSLAAK-------DGIDWIEYKRSLKKAEQWNVEVY 1172
>CA921657 similar to PIR|T03826|T03 cytochrome b245 beta chain homolog
RbohAp108 - Arabidopsis thaliana, partial (26%)
Length = 749
Score = 28.5 bits (62), Expect = 6.3
Identities = 30/112 (26%), Positives = 40/112 (34%), Gaps = 10/112 (8%)
Frame = -2
Query: 267 PLEWLIQLVPMLKKREFSISSSQSSHPNQVHLTVSVVSWTTPYKRKKKGLCS------SW 320
P EW FSI+S+ VH+ + WT KR +C S
Sbjct: 394 PFEW----------HPFSITSAPGDDYLSVHIR-QLGDWTQELKRVFSEVCEPPVSGRSG 248
Query: 321 LAALDPRDAVSLPVWFQKGSLPTPSPSLP----LILVGPGTGCAPFRGFIEE 368
L D SLP G P+ L+LVG G G PF +++
Sbjct: 247 LLRADETTKKSLPKLKIDGPYGAPAQDYRKYDVLLLVGLGIGATPFISILKD 92
>TC80501 similar to PIR|T52458|T52458 chlorophyll b synthase [imported] -
Arabidopsis thaliana, partial (59%)
Length = 1172
Score = 28.1 bits (61), Expect = 8.3
Identities = 26/87 (29%), Positives = 39/87 (43%), Gaps = 1/87 (1%)
Frame = -3
Query: 87 VSHSSDTGSVRSMHPG-KSSSNRNGYPDCFLKLVKNLPLTKPNSGKDVRHFEFEFVSHAI 145
++H S S+ +M KS+ GY F KL +G D F F+ ++
Sbjct: 798 ITHGSSKHSIGTMESSFKSAEKATGYQK-FFKLG-----WYD*TGPDTFKLFFGFLLLSV 637
Query: 146 EYDTGDILEVLPGQDSAAVDAFIRRCN 172
D+LEVL G+D +A A + CN
Sbjct: 636 ---CTDLLEVLGGEDVSATSAVLLSCN 565
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,284,983
Number of Sequences: 36976
Number of extensions: 260027
Number of successful extensions: 1234
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1211
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1223
length of query: 504
length of database: 9,014,727
effective HSP length: 100
effective length of query: 404
effective length of database: 5,317,127
effective search space: 2148119308
effective search space used: 2148119308
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148995.5


