

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.13 - phase: 0
(160 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
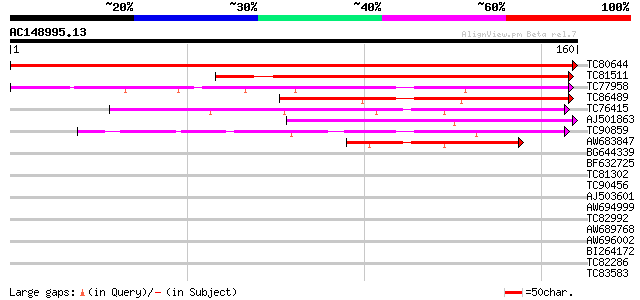
Score E
Sequences producing significant alignments: (bits) Value
TC80644 similar to GP|20259627|gb|AAM14170.1 unknown protein {Ar... 325 4e-90
TC81511 similar to GP|20259627|gb|AAM14170.1 unknown protein {Ar... 106 5e-24
TC77958 similar to GP|21592602|gb|AAM64551.1 unknown {Arabidopsi... 93 4e-20
TC86489 similar to GP|21592602|gb|AAM64551.1 unknown {Arabidopsi... 88 2e-18
TC76415 similar to PIR|T05479|T05479 hypothetical protein T8O5.1... 68 1e-12
AJ501863 similar to GP|21537049|gb| unknown {Arabidopsis thalian... 61 2e-10
TC90859 similar to GP|3337365|gb|AAC27410.1| expressed protein {... 54 3e-08
AW683847 weakly similar to GP|21537049|gb| unknown {Arabidopsis ... 47 3e-06
BG644339 weakly similar to PIR|T48401|T484 histone deacetylase-l... 30 0.32
BF632725 weakly similar to GP|21618118|gb unknown {Arabidopsis t... 28 2.1
TC81302 similar to GP|15529202|gb|AAK97695.1 AT3g01160/T4P13_15 ... 28 2.1
TC90456 weakly similar to PIR|G86466|G86466 hypothetical protein... 27 2.7
AJ503601 27 2.7
AW694999 similar to PIR|T02406|T024 hypothetical protein At2g445... 27 2.7
TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D... 27 3.6
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 27 3.6
AW696002 similar to GP|9294564|dbj| starch-branching enzyme-like... 27 4.6
BI264172 homologue to PIR|T45739|T457 transcription factor-like ... 27 4.6
TC82286 weakly similar to PIR|B96544|B96544 hypothetical protein... 27 4.6
TC83583 similar to PIR|T06379|T06379 SAR DNA-binding protein 2 -... 27 4.6
>TC80644 similar to GP|20259627|gb|AAM14170.1 unknown protein {Arabidopsis
thaliana}, partial (52%)
Length = 774
Score = 325 bits (834), Expect = 4e-90
Identities = 160/160 (100%), Positives = 160/160 (100%)
Frame = +2
Query: 1 MASSRKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNIEPKKGVPGLK 60
MASSRKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNIEPKKGVPGLK
Sbjct: 107 MASSRKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNIEPKKGVPGLK 286
Query: 61 RVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHE 120
RVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHE
Sbjct: 287 RVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHE 466
Query: 121 YLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
YLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED
Sbjct: 467 YLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 586
>TC81511 similar to GP|20259627|gb|AAM14170.1 unknown protein {Arabidopsis
thaliana}, partial (41%)
Length = 725
Score = 106 bits (264), Expect = 5e-24
Identities = 56/101 (55%), Positives = 67/101 (65%)
Frame = +1
Query: 59 LKRVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPP 118
L R KM+A +A +S+P+ IPDWSKIL +EYK + +E D D LPP
Sbjct: 298 LHRSRSKMDAG-----VAPASMPVKIPDWSKILGDEYKNNYTKRNYVEEEDEDDDEWLPP 462
Query: 119 HEYLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFE 159
HE+LARTR AS SVHEG GRTLKGRDL +RNAIW K GF+
Sbjct: 463 HEFLARTRVASFSVHEGVGRTLKGRDLSRLRNAIWAKTGFQ 585
>TC77958 similar to GP|21592602|gb|AAM64551.1 unknown {Arabidopsis
thaliana}, partial (39%)
Length = 787
Score = 93.2 bits (230), Expect = 4e-20
Identities = 68/183 (37%), Positives = 96/183 (52%), Gaps = 24/183 (13%)
Frame = +1
Query: 1 MASSRKSFLSRTSYIFPETNFNQKSSQGKEL-EFDEADVWNMSYSNS-NTNIEPKKGVPG 58
MA RK +R + TN+ Q S+ E+ +F E DVW++ N N P G
Sbjct: 145 MAKGRKFTTNRNERLLG-TNYIQSSNTTHEVTDFREEDVWSIVEDRDHNVNFTP--GDWD 315
Query: 59 LKRVSRK-------MEANNKVNPLASSS--------------LPMNIPDWSKILKEEYKK 97
+ SR+ + N V+ +A++S P+N+PDWSKIL+ E +
Sbjct: 316 SRAPSRRFNHGGLSLPFENPVSNVATTSSRNVHHQYRNVATSAPVNVPDWSKILRVESVE 495
Query: 98 KKESSDDEDEGDYDGVVQLPPHEYLARTRG-ASLSVHEGKGRTLKGRDLRSVRNAIWKKV 156
DD++E + +PPHEYLAR R A+ SV EG GRTLKGRDLR VR+A+W +
Sbjct: 496 SLHDMDDDNESE-----MVPPHEYLARGRKMAANSVFEGVGRTLKGRDLRRVRDAVWNQT 660
Query: 157 GFE 159
GF+
Sbjct: 661 GFD 669
>TC86489 similar to GP|21592602|gb|AAM64551.1 unknown {Arabidopsis
thaliana}, partial (41%)
Length = 1172
Score = 87.8 bits (216), Expect = 2e-18
Identities = 45/86 (52%), Positives = 62/86 (71%), Gaps = 3/86 (3%)
Frame = +2
Query: 77 SSSLPMNIPDWSKILKEEYKKK--KESSDDEDEGDYDGVVQLPPHEYLARTR-GASLSVH 133
++SLP+N+PDWSKIL+ + + +S DD+DE + +PPHEYLAR+R A+ SV
Sbjct: 548 ATSLPVNVPDWSKILRVDSVESLHDDSFDDDDESE-----MVPPHEYLARSRKNAAKSVF 712
Query: 134 EGKGRTLKGRDLRSVRNAIWKKVGFE 159
EG GRTLKGRDL VR+A+W + GF+
Sbjct: 713 EGVGRTLKGRDLSRVRDAVWSQTGFD 790
>TC76415 similar to PIR|T05479|T05479 hypothetical protein T8O5.180 -
Arabidopsis thaliana, partial (30%)
Length = 1114
Score = 68.2 bits (165), Expect = 1e-12
Identities = 53/180 (29%), Positives = 79/180 (43%), Gaps = 50/180 (27%)
Frame = +1
Query: 29 KELEFDEADVWNMSYSNSNTNIEPKKG-----------------VPGLKRVSRKMEANNK 71
++ +F+E ++W++ N E K +P + S AN
Sbjct: 280 RDEDFEEEEIWDVVKERPNYISEVHKPKEKKELSSLPIPTAARMIPIPRTSSGSSSANLS 459
Query: 72 VNPLA-SSSLPMNIPDWSKILKEEYKKKKESS---------------------------- 102
+A S P+NIPDWSKI K+ +S+
Sbjct: 460 HETMAFQQSAPVNIPDWSKIYGNNNNKQNKSTKKVSRYNEYGYYEGDDEVVYHGGEDGEE 639
Query: 103 DDEDEGDYDGVVQLPPHEY----LARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGF 158
DD+D+ +Y ++PPHE LAR++ +S SV EG GRTLKGRDL +RN++ K GF
Sbjct: 640 DDDDDDEYS--TRVPPHEIISRRLARSQISSFSVFEGVGRTLKGRDLSKMRNSVLIKTGF 813
>AJ501863 similar to GP|21537049|gb| unknown {Arabidopsis thaliana}, partial
(22%)
Length = 413
Score = 60.8 bits (146), Expect = 2e-10
Identities = 34/86 (39%), Positives = 48/86 (55%), Gaps = 4/86 (4%)
Frame = +2
Query: 79 SLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEYLAR----TRGASLSVHE 134
S PMN+P S + ++ +E + + +PPHEYL+R + S S+ E
Sbjct: 62 SAPMNVPMMSSAMANRARRYEEEDAHALNEEEEFRSTMPPHEYLSRQVDFSPMHSCSLFE 241
Query: 135 GKGRTLKGRDLRSVRNAIWKKVGFED 160
G GRTLKGRD+R VRNA+ + GF D
Sbjct: 242 GVGRTLKGRDMRQVRNAVLSQTGFFD 319
>TC90859 similar to GP|3337365|gb|AAC27410.1| expressed protein {Arabidopsis
thaliana}, partial (25%)
Length = 635
Score = 53.9 bits (128), Expect = 3e-08
Identities = 45/145 (31%), Positives = 75/145 (51%), Gaps = 6/145 (4%)
Frame = +3
Query: 20 NFNQKSSQGKELEFDEADVWNMSYSNSNTNIEPKKGVPGLKRVSRKMEANNKVNPLASS- 78
NF + S+ EF E++V + + + E KK + LK+ ++ ++++++ A +
Sbjct: 27 NFTKNMSE----EFLESEVLFFNQIHQS-EAEDKKVLMKLKK--EEINSDDEIDQAADNK 185
Query: 79 ---SLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTRGASL--SVH 133
S+PMNIP +++ +DEDE + +PPH +AR + S+
Sbjct: 186 MVRSMPMNIPSEGIF----HRRGNGYEEDEDEEE-----MVPPHLIMARRLAGKMTFSMC 338
Query: 134 EGKGRTLKGRDLRSVRNAIWKKVGF 158
G GRTLKGRDL VRN+I + GF
Sbjct: 339 SGHGRTLKGRDLSRVRNSILRMTGF 413
>AW683847 weakly similar to GP|21537049|gb| unknown {Arabidopsis thaliana},
partial (27%)
Length = 587
Score = 47.0 bits (110), Expect = 3e-06
Identities = 29/60 (48%), Positives = 38/60 (63%), Gaps = 10/60 (16%)
Frame = +1
Query: 96 KKKKE------SSDDEDEGDYDGVVQLPPHEY----LARTRGASLSVHEGKGRTLKGRDL 145
KKKK+ S DD+D+ +Y ++PPHE LAR++ +S SV EG GRTL GRDL
Sbjct: 217 KKKKKXTRD*FSXDDDDDDEYS--TRVPPHEIISRRLARSQISSFSVFEGVGRTLXGRDL 390
>BG644339 weakly similar to PIR|T48401|T484 histone deacetylase-like protein
- Arabidopsis thaliana, partial (26%)
Length = 763
Score = 30.4 bits (67), Expect = 0.32
Identities = 21/60 (35%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Frame = +1
Query: 59 LKRVSRKMEANNK-VNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLP 117
L+ ++ K+EA+ K V P A S P+ +KI KKE+ DED+ D +G +P
Sbjct: 370 LEALNGKLEADVKDVKPDAKKSAPVKDEKNAKIAAS----KKEAESDEDDSDSEGDSDVP 537
>BF632725 weakly similar to GP|21618118|gb unknown {Arabidopsis thaliana},
partial (53%)
Length = 621
Score = 27.7 bits (60), Expect = 2.1
Identities = 11/23 (47%), Positives = 17/23 (73%), Gaps = 1/23 (4%)
Frame = +1
Query: 72 VNPLASS-SLPMNIPDWSKILKE 93
VNPL+SS S P +PDW+++ +
Sbjct: 52 VNPLSSSFSTPAQVPDWNQVFAD 120
>TC81302 similar to GP|15529202|gb|AAK97695.1 AT3g01160/T4P13_15
{Arabidopsis thaliana}, partial (11%)
Length = 674
Score = 27.7 bits (60), Expect = 2.1
Identities = 25/93 (26%), Positives = 41/93 (43%), Gaps = 1/93 (1%)
Frame = +1
Query: 44 SNSNTNIEPKKGVPGLKRVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKK-ESS 102
S N K +P V ++ + +K N S ++ + K+ +E+ KKKK E S
Sbjct: 325 SRFNRMFTHKSFLPSSAPVDKRGKPKDKAN-----SQEHSLRHYYKMDEEDDKKKKVEQS 489
Query: 103 DDEDEGDYDGVVQLPPHEYLARTRGASLSVHEG 135
DE++ D + E L + + A L V G
Sbjct: 490 SDEEDNDDE-------EEELVKVKKAKLEVESG 567
>TC90456 weakly similar to PIR|G86466|G86466 hypothetical protein AAD39608.1
[imported] - Arabidopsis thaliana, partial (2%)
Length = 1332
Score = 27.3 bits (59), Expect = 2.7
Identities = 15/41 (36%), Positives = 22/41 (53%), Gaps = 2/41 (4%)
Frame = -2
Query: 15 IFPETNFNQK-SSQGKELE-FDEADVWNMSYSNSNTNIEPK 53
+ P FNQK Q E E F+ + +W +SNSN ++ K
Sbjct: 395 LIPHCRFNQKIPHQSVETEKFESSPIWITFHSNSNLDVNLK 273
>AJ503601
Length = 541
Score = 27.3 bits (59), Expect = 2.7
Identities = 16/57 (28%), Positives = 28/57 (49%)
Frame = +2
Query: 59 LKRVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQ 115
L R++ + N + + SS P + K +++ E S+ D G++DGVVQ
Sbjct: 323 LARMASVSKPNRRSKSIESSEKPTRSSTSNGSSKHTLEERPEGSESSD-GEFDGVVQ 490
>AW694999 similar to PIR|T02406|T024 hypothetical protein At2g44510
[imported] - Arabidopsis thaliana, partial (19%)
Length = 574
Score = 27.3 bits (59), Expect = 2.7
Identities = 16/57 (28%), Positives = 28/57 (49%)
Frame = +2
Query: 59 LKRVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQ 115
L R++ + N + + SS P + K +++ E S+ D G++DGVVQ
Sbjct: 149 LARMASVSKPNRRSKSIESSEKPTRSSTSNGSSKHTLEERPEGSESSD-GEFDGVVQ 316
>TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (7%)
Length = 354
Score = 26.9 bits (58), Expect = 3.6
Identities = 17/67 (25%), Positives = 31/67 (45%)
Frame = +1
Query: 92 KEEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTRGASLSVHEGKGRTLKGRDLRSVRNA 151
+EEY++++E +D+D+G G V P L + G E ++ N+
Sbjct: 103 EEEYEEEEEEEEDDDDG--SGAVYFPEAGDLEPSEGMEFESEEA---------AKAFYNS 249
Query: 152 IWKKVGF 158
++VGF
Sbjct: 250 YARRVGF 270
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 26.9 bits (58), Expect = 3.6
Identities = 14/47 (29%), Positives = 24/47 (50%)
Frame = +1
Query: 74 PLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHE 120
P +LP++ P W + +K EYK ++ + +V LPPH+
Sbjct: 58 PQDCENLPLSDPRWLQAMKTEYKALIDNKTWD-------LVPLPPHK 177
>AW696002 similar to GP|9294564|dbj| starch-branching enzyme-like protein
{Arabidopsis thaliana}, partial (8%)
Length = 654
Score = 26.6 bits (57), Expect = 4.6
Identities = 13/38 (34%), Positives = 24/38 (62%)
Frame = +1
Query: 92 KEEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTRGAS 129
+++YKKKKE +++ +E D++ P +L + RG S
Sbjct: 259 QKQYKKKKELNNNANETDFEAEKGYDPVGFLVK-RGVS 369
>BI264172 homologue to PIR|T45739|T457 transcription factor-like protein -
Arabidopsis thaliana, partial (4%)
Length = 568
Score = 26.6 bits (57), Expect = 4.6
Identities = 13/26 (50%), Positives = 15/26 (57%)
Frame = +1
Query: 4 SRKSFLSRTSYIFPETNFNQKSSQGK 29
S K FLS T IFP NF+ S+ K
Sbjct: 256 SEKIFLSTTCLIFPTVNFSMVLSEKK 333
>TC82286 weakly similar to PIR|B96544|B96544 hypothetical protein F4M15.4
[imported] - Arabidopsis thaliana, partial (5%)
Length = 1782
Score = 26.6 bits (57), Expect = 4.6
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = +2
Query: 81 PMNIPDWSKILKEEYKKKKESSDDEDEGDYDG 112
P I DW KEE +++ E +DE E DG
Sbjct: 389 PEFITDWGDEEKEEEEEENEEKEDEAEHMNDG 484
>TC83583 similar to PIR|T06379|T06379 SAR DNA-binding protein 2 - garden
pea, partial (33%)
Length = 852
Score = 26.6 bits (57), Expect = 4.6
Identities = 23/94 (24%), Positives = 44/94 (46%), Gaps = 3/94 (3%)
Frame = +2
Query: 53 KKGVPGLKRVSRKME--ANNKVNPLASSSLPMNIPDWSKI-LKEEYKKKKESSDDEDEGD 109
KKG GL ++ A++ + P+++S++ + P+ K+E K+KKE E + +
Sbjct: 278 KKGAGGLITPAKTYNTAADSVIEPMSNSAMDEDTPEPPVTDKKKEKKEKKEKKKKEKKEE 457
Query: 110 YDGVVQLPPHEYLARTRGASLSVHEGKGRTLKGR 143
V+ P E + + + KGR + R
Sbjct: 458 EVEDVEEPEEEVVKKEKKKKKKDST*KGRGAEWR 559
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.308 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,607,362
Number of Sequences: 36976
Number of extensions: 54657
Number of successful extensions: 349
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 333
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 339
length of query: 160
length of database: 9,014,727
effective HSP length: 88
effective length of query: 72
effective length of database: 5,760,839
effective search space: 414780408
effective search space used: 414780408
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC148995.13


