

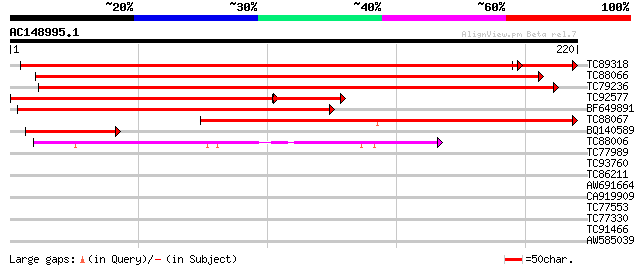
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.1 + phase: 0
(220 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89318 homologue to GP|21805708|gb|AAM76759.1 hypothetical prot... 380 e-112
TC88066 similar to GP|21805708|gb|AAM76759.1 hypothetical protei... 323 2e-89
TC79236 similar to PIR|T48554|T48554 lysine decarboxylase-like p... 288 1e-78
TC92577 homologue to GP|21805708|gb|AAM76759.1 hypothetical prot... 206 6e-65
BF649891 homologue to GP|13605607|gb T3B23.2/T3B23.2 {Arabidopsi... 220 4e-58
TC88067 similar to GP|20197602|gb|AAM15149.1 expressed protein {... 214 3e-56
BQ140589 homologue to GP|13605607|gb| T3B23.2/T3B23.2 {Arabidops... 64 3e-11
TC88006 similar to GP|16649027|gb|AAL24365.1 unknown protein {Ar... 49 2e-06
TC77989 similar to GP|16904543|emb|CAD10740. lipoxygenase {Coryl... 29 1.2
TC93760 similar to PIR|T48153|T48153 hypothetical protein T10O8.... 29 1.6
TC86211 homologue to GP|13194621|gb|AAK15493.1 brassinosteroid b... 29 1.6
AW691664 similar to PIR|T52061|T52 tRNA isopentenyltransferase (... 28 2.8
CA919909 28 3.6
TC77553 homologue to GP|1066499|gb|AAB41904.1| NADH-dependent gl... 27 4.7
TC77330 similar to GP|13161397|dbj|BAB33033. CPRD2 {Vigna unguic... 27 6.2
TC91466 similar to GP|16930705|gb|AAL32018.1 AT5g35180/T25C13_60... 27 8.0
AW585039 similar to GP|13605859|gb AT4g02590/T10P11_13 {Arabidop... 27 8.0
>TC89318 homologue to GP|21805708|gb|AAM76759.1 hypothetical protein
{Arabidopsis thaliana}, partial (87%)
Length = 1078
Score = 380 bits (976), Expect(2) = e-112
Identities = 186/195 (95%), Positives = 194/195 (99%)
Frame = +1
Query: 5 NGEIKLSKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQA 64
NGEI++SKFKR+CVFCGSSPGKK++YQDAA+ LGNELVSRNIDLVYGGGSIGLMGLVSQA
Sbjct: 76 NGEIRVSKFKRVCVFCGSSPGKKSSYQDAAIELGNELVSRNIDLVYGGGSIGLMGLVSQA 255
Query: 65 VHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLE 124
VHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLE
Sbjct: 256 VHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLE 435
Query: 125 ELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKEL 184
ELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPT+KEL
Sbjct: 436 ELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTSKEL 615
Query: 185 VKKLEEYVPCHEGVA 199
VKKLE+YVPCHEGVA
Sbjct: 616 VKKLEDYVPCHEGVA 660
Score = 42.4 bits (98), Expect(2) = e-112
Identities = 17/25 (68%), Positives = 23/25 (92%)
Frame = +2
Query: 196 EGVASKLSWQMEQQLAYPQDYDISR 220
+ + SKLSWQMEQ+LAYP++Y+ISR
Sbjct: 650 KALLSKLSWQMEQELAYPEEYNISR 724
>TC88066 similar to GP|21805708|gb|AAM76759.1 hypothetical protein
{Arabidopsis thaliana}, partial (93%)
Length = 922
Score = 323 bits (829), Expect = 2e-89
Identities = 155/197 (78%), Positives = 181/197 (91%)
Frame = +2
Query: 11 SKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDGGR 70
S+FKRICV+CGS+PGK +YQ AA+ LG +LV RNIDLVYGGGSIGLMG +SQ V+DGGR
Sbjct: 227 SRFKRICVYCGSTPGKNPSYQIAAIQLGKQLVERNIDLVYGGGSIGLMGRISQVVYDGGR 406
Query: 71 HVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLEELLEVI 130
HV+GVIPKTLM RE+TGETVGEV+AV+DMHQRKAEMA+ +DAFIALPGGYGTLEELLE+I
Sbjct: 407 HVLGVIPKTLMLREITGETVGEVRAVSDMHQRKAEMARQADAFIALPGGYGTLEELLEII 586
Query: 131 TWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEE 190
TWAQLGIHDKPVGL+NVDGY+NSLL+F+DKAV+EGF++P ARHIIVSA TA++L+ KLEE
Sbjct: 587 TWAQLGIHDKPVGLLNVDGYYNSLLAFMDKAVDEGFVTPAARHIIVSAHTAQDLMCKLEE 766
Query: 191 YVPCHEGVASKLSWQME 207
YVP H GVA KLSW+ME
Sbjct: 767 YVPKHCGVAPKLSWEME 817
>TC79236 similar to PIR|T48554|T48554 lysine decarboxylase-like protein -
Arabidopsis thaliana, partial (93%)
Length = 1142
Score = 288 bits (737), Expect = 1e-78
Identities = 136/202 (67%), Positives = 167/202 (82%)
Frame = +1
Query: 12 KFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDGGRH 71
KFKR+CVFCGS+ G + + DAA+ L +ELV RNIDLVYGGGS+GLMGL+SQ +++GG H
Sbjct: 40 KFKRVCVFCGSNSGNRQVFSDAAIQLADELVKRNIDLVYGGGSVGLMGLISQKMYNGGCH 219
Query: 72 VIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLEELLEVIT 131
V+GVIPK LMP E++GE VGEV+ V+DMH+RKA MA+ ++AFIALPGGYGT+EELLE+IT
Sbjct: 220 VLGVIPKALMPHEISGEAVGEVRIVSDMHERKAAMAQEAEAFIALPGGYGTMEELLEMIT 399
Query: 132 WAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEEY 191
WAQLGIH KPVGL+NVDGY+NSLL+ D VEEGFI P+AR I+VSA +AKEL+ K+E Y
Sbjct: 400 WAQLGIHKKPVGLLNVDGYYNSLLALFDNGVEEGFIKPSARSIVVSASSAKELMLKMESY 579
Query: 192 VPCHEGVASKLSWQMEQQLAYP 213
P HE VA SWQM+Q YP
Sbjct: 580 SPSHEHVAPHESWQMKQLGNYP 645
>TC92577 homologue to GP|21805708|gb|AAM76759.1 hypothetical protein
{Arabidopsis thaliana}, partial (55%)
Length = 511
Score = 206 bits (524), Expect(2) = 6e-65
Identities = 102/104 (98%), Positives = 103/104 (98%)
Frame = +3
Query: 1 MDSRNGEIKLSKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGL 60
MDSRNGEIKLSKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGL
Sbjct: 120 MDSRNGEIKLSKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGL 299
Query: 61 VSQAVHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKA 104
VSQAVHDGGRHVIGVIPKTLMPRELTGETV EVKAVADMHQRK+
Sbjct: 300 VSQAVHDGGRHVIGVIPKTLMPRELTGETVREVKAVADMHQRKS 431
Score = 58.5 bits (140), Expect(2) = 6e-65
Identities = 28/28 (100%), Positives = 28/28 (100%)
Frame = +1
Query: 103 KAEMAKHSDAFIALPGGYGTLEELLEVI 130
KAEMAKHSDAFIALPGGYGTLEELLEVI
Sbjct: 427 KAEMAKHSDAFIALPGGYGTLEELLEVI 510
>BF649891 homologue to GP|13605607|gb T3B23.2/T3B23.2 {Arabidopsis thaliana},
partial (54%)
Length = 382
Score = 220 bits (560), Expect = 4e-58
Identities = 108/123 (87%), Positives = 117/123 (94%)
Frame = +1
Query: 4 RNGEIKLSKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQ 63
R E+K+SKFKRICVFCGSSPG KT+Y+DAA+ LG ELVSRNIDLVYGGGSIGLMGL+SQ
Sbjct: 13 REIEMKVSKFKRICVFCGSSPGNKTSYKDAAIELGKELVSRNIDLVYGGGSIGLMGLISQ 192
Query: 64 AVHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTL 123
AV+DGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEM+ +SDAFIALPGGYGTL
Sbjct: 193 AVYDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMSXNSDAFIALPGGYGTL 372
Query: 124 EEL 126
EEL
Sbjct: 373 EEL 381
>TC88067 similar to GP|20197602|gb|AAM15149.1 expressed protein {Arabidopsis
thaliana}, partial (62%)
Length = 882
Score = 214 bits (544), Expect = 3e-56
Identities = 115/187 (61%), Positives = 132/187 (70%), Gaps = 41/187 (21%)
Frame = +1
Query: 75 VIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLEELLEVITWAQ 134
VIPKTLM RE+TGETVGEV+AV+DMHQRKAEMA+ +DAFIALPGGYGTLEELLE+ITWAQ
Sbjct: 1 VIPKTLMLREITGETVGEVRAVSDMHQRKAEMARQADAFIALPGGYGTLEELLEIITWAQ 180
Query: 135 LGIHDKP-----------------------------------------VGLVNVDGYFNS 153
LGIHDKP VGL+NVDGY+NS
Sbjct: 181 LGIHDKPVRLLPFV*RRSQSKNDITYLMYITIKMRIKCKIGNNEWDA*VGLLNVDGYYNS 360
Query: 154 LLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEEYVPCHEGVASKLSWQMEQQLAYP 213
LL+F+DKAV+EGF++P ARHIIVSA TA++L+ KLEEYVP H GVA KLSW+MEQQL
Sbjct: 361 LLAFMDKAVDEGFVTPAARHIIVSAHTAQDLMCKLEEYVPKHCGVAPKLSWEMEQQLVNT 540
Query: 214 QDYDISR 220
DISR
Sbjct: 541 AKLDISR 561
>BQ140589 homologue to GP|13605607|gb| T3B23.2/T3B23.2 {Arabidopsis
thaliana}, partial (15%)
Length = 118
Score = 64.3 bits (155), Expect = 3e-11
Identities = 29/37 (78%), Positives = 34/37 (91%)
Frame = +1
Query: 7 EIKLSKFKRICVFCGSSPGKKTTYQDAAMNLGNELVS 43
E+K SKFKRICVFCGSSPG K++Y+DAA+ LGNELVS
Sbjct: 7 EMKESKFKRICVFCGSSPGNKSSYKDAAIELGNELVS 117
>TC88006 similar to GP|16649027|gb|AAL24365.1 unknown protein {Arabidopsis
thaliana}, partial (65%)
Length = 1153
Score = 48.5 bits (114), Expect = 2e-06
Identities = 53/183 (28%), Positives = 77/183 (41%), Gaps = 24/183 (13%)
Frame = +1
Query: 10 LSKFKRICVFCGSSP-GKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDG 68
+++ R V+ GSS G ++ A L E+ + + G GLM V+Q
Sbjct: 304 INRLGRGIVYLGSSRMGSSHSHYVQAQELAKEIANLLDSTTWSGAGPGLMDAVTQGALLA 483
Query: 69 GRHVIGV-----------------IPKT--LMPRELTGETVGEVKAVADMHQRKAEMAKH 109
G+ V G +P L R + G V AV R K
Sbjct: 484 GKPVGGFKIGREAGEWTASNFHPYLPSENYLTCRFFSARKHGLVDAVV----RNNSFDK- 648
Query: 110 SDAFIALPGGYGTLEELLEVITWAQL-GIHDK---PVGLVNVDGYFNSLLSFIDKAVEEG 165
A +ALPGG GTL+EL E++ QL I K P L+N D +++ LL F+D + G
Sbjct: 649 -TAVVALPGGIGTLDELFEMLALIQLERIGSKLPVPFLLMNYDSFYSKLLDFLDVCEDRG 825
Query: 166 FIS 168
+S
Sbjct: 826 TVS 834
>TC77989 similar to GP|16904543|emb|CAD10740. lipoxygenase {Corylus
avellana}, partial (54%)
Length = 1763
Score = 29.3 bits (64), Expect = 1.2
Identities = 25/100 (25%), Positives = 42/100 (42%), Gaps = 6/100 (6%)
Frame = +3
Query: 122 TLEELLEVITWAQLGIHD------KPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHII 175
T++E LE L HD + N Y L F+ ++G + P A +
Sbjct: 105 TIDEALESDKLYILDHHDALMPYLSRINSTNTKTYATRTLLFLQ---DDGTLKPLAIELS 275
Query: 176 VSAPTAKELVKKLEEYVPCHEGVASKLSWQMEQQLAYPQD 215
+ P ++ + + P HEGVA+ + WQ+ + A D
Sbjct: 276 LPHPQGEQHGAVSKVFTPSHEGVAATV-WQLAKAYAAVND 392
>TC93760 similar to PIR|T48153|T48153 hypothetical protein T10O8.20 -
Arabidopsis thaliana, partial (18%)
Length = 835
Score = 28.9 bits (63), Expect = 1.6
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = -3
Query: 189 EEYVPCHEGVASKLSWQMEQQ 209
+EYVP G+A K SWQ++QQ
Sbjct: 770 QEYVPKDCGMAPKRSWQIDQQ 708
>TC86211 homologue to GP|13194621|gb|AAK15493.1 brassinosteroid biosynthetic
protein LKB {Pisum sativum}, partial (60%)
Length = 1141
Score = 28.9 bits (63), Expect = 1.6
Identities = 25/89 (28%), Positives = 38/89 (42%)
Frame = +1
Query: 35 MNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDGGRHVIGVIPKTLMPRELTGETVGEVK 94
MNL +V+ DL GG L++ +G H G+ T++ E+ V+
Sbjct: 526 MNLALAVVAELDDLTVGG-------LINGYGIEGSSHKYGLFSDTVVAFEIVLADGSLVR 684
Query: 95 AVADMHQRKAEMAKHSDAFIALPGGYGTL 123
A D ++SD F A+P GTL
Sbjct: 685 ATKDN--------EYSDLFYAIPWSQGTL 747
>AW691664 similar to PIR|T52061|T52 tRNA isopentenyltransferase (EC 2.5.1.8)
[imported] - Arabidopsis thaliana, partial (32%)
Length = 665
Score = 28.1 bits (61), Expect = 2.8
Identities = 14/40 (35%), Positives = 28/40 (70%), Gaps = 1/40 (2%)
Frame = +3
Query: 29 TYQDAAMNLGNELVSRN-IDLVYGGGSIGLMGLVSQAVHD 67
T++D+A+ + +E++SRN + ++ GG + + LVSQ + D
Sbjct: 279 TFRDSAIPIIDEILSRNHLPVIVGGTNYYIQALVSQFLLD 398
>CA919909
Length = 788
Score = 27.7 bits (60), Expect = 3.6
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = +3
Query: 163 EEGFISPNARHIIVSAPTAKELV 185
+ G SPN +HI ++A T KEL+
Sbjct: 513 KRGICSPN*KHISIAASTTKELI 581
>TC77553 homologue to GP|1066499|gb|AAB41904.1| NADH-dependent glutamate
synthase {Medicago sativa}, partial (37%)
Length = 3067
Score = 27.3 bits (59), Expect = 4.7
Identities = 14/35 (40%), Positives = 21/35 (60%)
Frame = +1
Query: 135 LGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISP 169
LGI + PV + N++ + IDKA EEG++ P
Sbjct: 1264 LGIIENPVSIKNIE------CAIIDKAFEEGWMIP 1350
>TC77330 similar to GP|13161397|dbj|BAB33033. CPRD2 {Vigna unguiculata},
partial (29%)
Length = 1295
Score = 26.9 bits (58), Expect = 6.2
Identities = 15/63 (23%), Positives = 31/63 (48%)
Frame = +1
Query: 106 MAKHSDAFIALPGGYGTLEELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEG 165
M K S++ I P YGTL ++ + W Q G + + + + ++ + F+ K+
Sbjct: 457 MDKISESEIPFPHRYGTLYKIQYAVHWHQEGDEVEKLHINWIRKLYSYMEPFVSKSPRAA 636
Query: 166 FIS 168
+I+
Sbjct: 637 YIN 645
>TC91466 similar to GP|16930705|gb|AAL32018.1 AT5g35180/T25C13_60
{Arabidopsis thaliana}, partial (21%)
Length = 682
Score = 26.6 bits (57), Expect = 8.0
Identities = 18/54 (33%), Positives = 24/54 (44%)
Frame = -2
Query: 131 TWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKEL 184
TW + K + N+D +NSLLS I + SP + I S PT L
Sbjct: 201 TWKHTFL*SKEIEDDNIDDQWNSLLSDIP---NRSYFSPILCYPICSTPTTMNL 49
>AW585039 similar to GP|13605859|gb AT4g02590/T10P11_13 {Arabidopsis
thaliana}, partial (46%)
Length = 628
Score = 26.6 bits (57), Expect = 8.0
Identities = 16/48 (33%), Positives = 26/48 (53%)
Frame = +2
Query: 167 ISPNARHIIVSAPTAKELVKKLEEYVPCHEGVASKLSWQMEQQLAYPQ 214
++PN+ I + T + + E +PCH +AS L + EQ+ A PQ
Sbjct: 41 LNPNSTFTINTNSTFRASNQIQRELLPCHNHLASVL--ECEQEEAKPQ 178
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,077,959
Number of Sequences: 36976
Number of extensions: 69929
Number of successful extensions: 340
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 339
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 340
length of query: 220
length of database: 9,014,727
effective HSP length: 92
effective length of query: 128
effective length of database: 5,612,935
effective search space: 718455680
effective search space used: 718455680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148995.1


