

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148994.4 - phase: 0
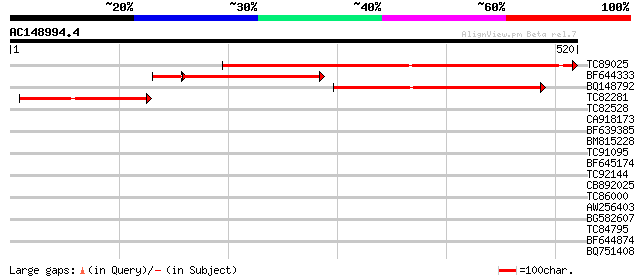
(520 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89025 similar to GP|2462911|emb|CAB06081.1 UDP-glucose:sterol ... 506 e-144
BF644333 similar to GP|2462911|emb UDP-glucose:sterol glucosyltr... 284 5e-88
BQ148792 similar to PIR|D96499|D96 probable UDP-glucose sterol g... 250 1e-66
TC82281 similar to GP|2462931|emb|CAB06082.1 UDP-glucose:sterol ... 137 1e-32
TC82528 weakly similar to PIR|C86356|C86356 hypothetical protein... 38 0.008
CA918173 weakly similar to GP|11034537|dbj putative glucosyl tra... 37 0.014
BF639385 weakly similar to GP|22830963|dbj putative UDP-glucose:... 37 0.024
BM815228 similar to GP|22136000|gb unknown protein {Arabidopsis ... 33 0.27
TC91095 weakly similar to GP|21435782|gb|AAM53963.1 UDP-glucosyl... 30 1.7
BF645174 similar to GP|19347728|gb putative receptor protein kin... 30 2.3
TC92144 weakly similar to GP|13620869|dbj|BAB41024. UDP-glucose:... 30 3.0
CB892025 similar to PIR|T06371|T06 probable UDP-glucuronosyltran... 30 3.0
TC86000 phosphoinositide-specific phospholipase C [Medicago trun... 29 5.0
AW256403 similar to PIR|F96828|F968 hypothetical protein F19K16.... 29 5.0
BG582607 28 8.6
TC84795 weakly similar to GP|14532546|gb|AAK64001.1 At1g22370/T1... 28 8.6
BF644874 GP|4521190|db Pib {Oryza sativa}, partial (0%) 28 8.6
BQ751408 weakly similar to GP|7297956|gb| bun gene product {alt ... 28 8.6
>TC89025 similar to GP|2462911|emb|CAB06081.1 UDP-glucose:sterol
glucosyltransferase {Avena sativa}, partial (47%)
Length = 1157
Score = 506 bits (1303), Expect = e-144
Identities = 236/325 (72%), Positives = 274/325 (83%)
Frame = +1
Query: 196 VPLHIFFTMPWTPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLINEFRKKKLKLR 255
+P+HIFFTMPWTPT+DFPHPLSRV+Q GYRLSYQIVD+LIWLGIRD+IN+ RKKKLKLR
Sbjct: 4 IPIHIFFTMPWTPTADFPHPLSRVKQQAGYRLSYQIVDSLIWLGIRDMINDLRKKKLKLR 183
Query: 256 AVTYLRGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDIVGFCFLDLASNYEPPKSLVDWL 315
VTYL GS F D+P+ YIWSPHLVPKPKDWGP ID+VGFCFLDLASNYEPP+SLV WL
Sbjct: 184 PVTYLSGSQGFENDIPHAYIWSPHLVPKPKDWGPKIDVVGFCFLDLASNYEPPESLVKWL 363
Query: 316 EEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYL 375
E+G+ PIY+GFGSLP+Q+P+KMT+IIV+ALE TGQRGIINKGWGGLG+L E S+YL
Sbjct: 364 EDGDKPIYIGFGSLPVQDPKKMTQIIVEALETTGQRGIINKGWGGLGDLTE--PKDSIYL 537
Query: 376 LDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAP 435
LDN PHDWLF +C AVVHHGGAGTTAAGL+A CPTT+VPFFGDQPFWGERVH RGVGP P
Sbjct: 538 LDNVPHDWLFLQCKAVVHHGGAGTTAAGLKAACPTTIVPFFGDQPFWGERVHDRGVGPPP 717
Query: 436 IRVEEFTLERLVDAIRFMLNPEVKKRAVELANAMKNEDGVAGAVNAFYKHYPREKPDTEA 495
I V+EF+L +L+DAI FML+P+VK+ A+ELA AM+NEDGV GAV AF+K P++KP+T
Sbjct: 718 IPVDEFSLPKLIDAINFMLDPKVKEHAIELAKAMENEDGVTGAVKAFFKQLPQKKPETNT 897
Query: 496 EPRPVPSVHKHLSIRGCFGCYHSSA 520
EP P +I CFG SSA
Sbjct: 898 EPSPSSCFS---NIARCFGHS*SSA 963
>BF644333 similar to GP|2462911|emb UDP-glucose:sterol glucosyltransferase
{Avena sativa}, partial (23%)
Length = 473
Score = 284 bits (726), Expect(2) = 5e-88
Identities = 130/130 (100%), Positives = 130/130 (100%)
Frame = +3
Query: 159 LPACNSRYPESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFTMPWTPTSDFPHPLSR 218
LPACNSRYPESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFTMPWTPTSDFPHPLSR
Sbjct: 84 LPACNSRYPESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFTMPWTPTSDFPHPLSR 263
Query: 219 VRQPIGYRLSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIWSP 278
VRQPIGYRLSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIWSP
Sbjct: 264 VRQPIGYRLSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIWSP 443
Query: 279 HLVPKPKDWG 288
HLVPKPKDWG
Sbjct: 444 HLVPKPKDWG 473
Score = 59.3 bits (142), Expect(2) = 5e-88
Identities = 29/31 (93%), Positives = 30/31 (96%)
Frame = +2
Query: 132 KNKGFLPSGPSEIHLQRSQIRAIIHSLLPAC 162
KNKGFLPSGPSEIHLQRSQIRAIIHS LP+C
Sbjct: 2 KNKGFLPSGPSEIHLQRSQIRAIIHS-LPSC 91
>BQ148792 similar to PIR|D96499|D96 probable UDP-glucose sterol
glucosyltransferase [imported] - Arabidopsis thaliana,
partial (31%)
Length = 701
Score = 250 bits (638), Expect = 1e-66
Identities = 110/194 (56%), Positives = 153/194 (78%)
Frame = +3
Query: 298 FLDLASNYEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKG 357
FL SNY+P + + W+++G P+Y GFGS+PL++P+ T +I++AL++T QRGII++G
Sbjct: 3 FLRHESNYQPREDFLHWIKKGPPPLYFGFGSMPLEDPKITTDVILKALKETEQRGIIDRG 182
Query: 358 WGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFG 417
WG LGNL E+ S +V+LL+ CPHDWLFP+C+AVVHHGGAGTTA GL++ CPTT+VPFFG
Sbjct: 183 WGNLGNLTEV--SDNVFLLEECPHDWLFPQCSAVVHHGGAGTTATGLKSGCPTTIVPFFG 356
Query: 418 DQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAMKNEDGVAG 477
DQ FWG+R+H + +GPAPI + E +E L +AI+FML PEVK R +E+A +++EDGVA
Sbjct: 357 DQFFWGDRIHQKELGPAPIPISELNVENLSNAIKFMLQPEVKSRTMEVAKLIESEDGVAA 536
Query: 478 AVNAFYKHYPREKP 491
V+AF++H P E P
Sbjct: 537 XVDAFHRHLPDELP 578
>TC82281 similar to GP|2462931|emb|CAB06082.1 UDP-glucose:sterol
glucosyltransferase {Arabidopsis thaliana}, partial
(18%)
Length = 799
Score = 137 bits (344), Expect = 1e-32
Identities = 77/122 (63%), Positives = 89/122 (72%), Gaps = 1/122 (0%)
Frame = +1
Query: 10 QLRWLKNAFMVKDDGTVEIDVPGNIKPLALENGTG-VIDPSDESCNETINEDIEPIRPQQ 68
+LR L VKDDGTVE DVP +++P AL G+ V + D+S T D++ I P
Sbjct: 442 KLRLLNRIATVKDDGTVEFDVPIDVEPDALGAGSKHVNNVIDDSHGAT---DLDYIPPLN 612
Query: 69 IAMLIVGTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGGDPKVLAE 128
I MLIVGTRGDVQPFVAIGKRLQ GHRVRL TH NF++FVL+AGLEFYPLGG PKVLA
Sbjct: 613 IVMLIVGTRGDVQPFVAIGKRLQDYGHRVRLXTHSNFKEFVLTAGLEFYPLGGXPKVLAG 792
Query: 129 YM 130
YM
Sbjct: 793 YM 798
>TC82528 weakly similar to PIR|C86356|C86356 hypothetical protein AAF87257.1
[imported] - Arabidopsis thaliana, partial (35%)
Length = 1376
Score = 38.1 bits (87), Expect = 0.008
Identities = 33/131 (25%), Positives = 55/131 (41%), Gaps = 11/131 (8%)
Frame = +1
Query: 301 LASN-YEPPKSLVDWLE--EGENPIYVGFGSLPLQEPEKMTRIIVQALEQTG------QR 351
L+SN ++ ++WLE E E+ +YV FGS+ + P ++ + +
Sbjct: 643 LSSNLWKEDTKCLEWLESKEPESVVYVNFGSITVMTPNQLLEFAWGLADSKKPFLWIIRP 822
Query: 352 GIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAECP 409
++ G L + E S + CP + + P + H G +T + A P
Sbjct: 823 DLVIGGSFILSSEFENEISDRGLITSWCPQEQVLIHPSIGGFLTHCGWNSTTESICAGVP 1002
Query: 410 TTVVPFFGDQP 420
PFFGDQP
Sbjct: 1003MLCWPFFGDQP 1035
>CA918173 weakly similar to GP|11034537|dbj putative glucosyl transferase
{Oryza sativa (japonica cultivar-group)}, partial (19%)
Length = 769
Score = 37.4 bits (85), Expect = 0.014
Identities = 35/121 (28%), Positives = 52/121 (42%), Gaps = 13/121 (10%)
Frame = +2
Query: 311 LVDWL--EEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGI----INKGWGGLGNL 364
L++WL +E E+ +YV FGSL ++ P I LE +G I NK G G L
Sbjct: 302 LLNWLNSKENESVLYVSFGSL-IRLPHAQLVEIAHGLENSGHNFIWVIKNNKDEDGEGFL 478
Query: 365 AEL-----NTSKSVYLLDNCPHDWL--FPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFG 417
E ++K + D P + +P +V H G +T + A P P F
Sbjct: 479 QEFEKRMKESNKGYIIWDWAPQLLILEYPAIGGIVTHCGWNSTLESVNAGLPMITWPVFA 658
Query: 418 D 418
+
Sbjct: 659 E 661
>BF639385 weakly similar to GP|22830963|dbj putative UDP-glucose:sterol
glucosyltransferase {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 554
Score = 36.6 bits (83), Expect = 0.024
Identities = 18/38 (47%), Positives = 25/38 (65%)
Frame = +2
Query: 10 QLRWLKNAFMVKDDGTVEIDVPGNIKPLALENGTGVID 47
+LR L VKDDGTVE DVP +++P AL G+ ++
Sbjct: 404 KLRLLNKIATVKDDGTVEFDVPIDVEPEALGAGSAHVN 517
>BM815228 similar to GP|22136000|gb unknown protein {Arabidopsis thaliana},
partial (43%)
Length = 705
Score = 33.1 bits (74), Expect = 0.27
Identities = 26/88 (29%), Positives = 40/88 (44%), Gaps = 9/88 (10%)
Frame = -1
Query: 92 ADGHRVRLATHKNFEDFVLSAGLEFYPLGGDPKVLAEYMVKNKGFLPSGPSEIHLQRS-- 149
AD H VR AT +F DF+L++ ++ G V+ N G+ PS+I Q +
Sbjct: 363 ADSHMVRNATRLSFNDFILAS---YFAFSGPLPVMTN---NNLGYKHCFPSDISFQMTSS 202
Query: 150 -------QIRAIIHSLLPACNSRYPESN 170
I A S + A + R P+S+
Sbjct: 201 SSESSSFNIAATRLSYIKAASKRMPKSS 118
>TC91095 weakly similar to GP|21435782|gb|AAM53963.1 UDP-glucosyltransferase
{Stevia rebaudiana}, partial (24%)
Length = 934
Score = 30.4 bits (67), Expect = 1.7
Identities = 22/87 (25%), Positives = 38/87 (43%), Gaps = 7/87 (8%)
Frame = +1
Query: 394 HGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEE---FTLERLVDAI 450
H G+G+ L ++PF DQ + + + VG +R EE FT + A+
Sbjct: 364 HCGSGSMIENLYFGHVLVMLPFLLDQALYSRVMQEKKVGIEIVRNEEDGSFTRNSVAKAL 543
Query: 451 RFML----NPEVKKRAVELANAMKNED 473
RF + + +K A E+ N++
Sbjct: 544 RFTMVDEEGSDYRKNAKEIGKKFSNKE 624
>BF645174 similar to GP|19347728|gb putative receptor protein kinase
{Arabidopsis thaliana}, partial (46%)
Length = 633
Score = 30.0 bits (66), Expect = 2.3
Identities = 28/79 (35%), Positives = 39/79 (48%), Gaps = 13/79 (16%)
Frame = +1
Query: 203 TMPWTPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLIN--------EFRKK---- 250
T+ T D + LSR + + ++ SYQ D ++ L IR L N FRK+
Sbjct: 145 TLEATRNFDEENVLSRGKHGLVFKASYQ--DGMV-LSIRRLPNGSTLMDEATFRKEAESL 315
Query: 251 -KLKLRAVTYLRGSYTFPP 268
K+K R +T LRG Y PP
Sbjct: 316 GKVKHRNLTVLRGYYAGPP 372
>TC92144 weakly similar to GP|13620869|dbj|BAB41024. UDP-glucose:flavonoid
3-O-glucosyltransferase {Vitis vinifera}, partial (34%)
Length = 1040
Score = 29.6 bits (65), Expect = 3.0
Identities = 13/40 (32%), Positives = 26/40 (64%), Gaps = 2/40 (5%)
Frame = +2
Query: 312 VDWLEEGENP--IYVGFGSLPLQEPEKMTRIIVQALEQTG 349
++WL++ EN +Y+ FGS+ P ++T + ++LE+ G
Sbjct: 812 LEWLDQHENSSVVYISFGSVVTPPPHELT-ALAESLEECG 928
>CB892025 similar to PIR|T06371|T06 probable UDP-glucuronosyltransferase (EC
2.4.1.-) - garden pea, partial (32%)
Length = 708
Score = 29.6 bits (65), Expect = 3.0
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 3/42 (7%)
Frame = +2
Query: 298 FLDLASN-YEPPKSLVDWLEEGE--NPIYVGFGSLPLQEPEK 336
F L SN ++ ++WLE E + +YV FGS+ + PEK
Sbjct: 449 FATLDSNLWKEDTKCLEWLESKEPASVVYVNFGSITIMSPEK 574
>TC86000 phosphoinositide-specific phospholipase C [Medicago truncatula]
Length = 2160
Score = 28.9 bits (63), Expect = 5.0
Identities = 19/56 (33%), Positives = 29/56 (50%), Gaps = 2/56 (3%)
Frame = +3
Query: 456 PEVKKRAVELANAMKNEDGV--AGAVNAFYKHYPREKPDTEAEPRPVPSVHKHLSI 509
PE+K EL + +E G+ A + +F +E+ TE E + + HKHLSI
Sbjct: 219 PEIK----ELYHRYSDEGGIMTASHLRSFLIEVQKEEKITEEETQAIIDGHKHLSI 374
>AW256403 similar to PIR|F96828|F968 hypothetical protein F19K16.26
[imported] - Arabidopsis thaliana, partial (15%)
Length = 499
Score = 28.9 bits (63), Expect = 5.0
Identities = 20/68 (29%), Positives = 31/68 (45%)
Frame = +3
Query: 227 LSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIWSPHLVPKPKD 286
L Y +++++ G LIN + + AV G +T PPDM SP KP+
Sbjct: 246 LIYVVLESIARFGCFQLINHWISPGIVAAAVDVAGGIFTVPPDMRDAVTRSPE---KPRG 416
Query: 287 WGPNIDIV 294
+ + D V
Sbjct: 417 FEEHDDEV 440
>BG582607
Length = 754
Score = 28.1 bits (61), Expect = 8.6
Identities = 17/46 (36%), Positives = 24/46 (51%)
Frame = -2
Query: 225 YRLSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDM 270
YRLS+ +V L+WL + ++ E + L L GSY F P M
Sbjct: 384 YRLSHYMVQNLLWLLLLNMKVEQEHQDLVWFQFHDLHGSYIFCPMM 247
>TC84795 weakly similar to GP|14532546|gb|AAK64001.1 At1g22370/T16E15_3
{Arabidopsis thaliana}, partial (33%)
Length = 605
Score = 28.1 bits (61), Expect = 8.6
Identities = 14/44 (31%), Positives = 20/44 (44%), Gaps = 2/44 (4%)
Frame = +1
Query: 379 CPHDWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQP 420
CP + + P + H G +T + A P PF+GDQP
Sbjct: 76 CPQEQVLNHPSIGGFLTHCGWNSTVESVLAGVPMLCWPFYGDQP 207
>BF644874 GP|4521190|db Pib {Oryza sativa}, partial (0%)
Length = 675
Score = 28.1 bits (61), Expect = 8.6
Identities = 11/36 (30%), Positives = 16/36 (43%)
Frame = +2
Query: 285 KDWGPNIDIVGFCFLDLASNYEPPKSLVDWLEEGEN 320
K W + V C D+ YE K + W++ EN
Sbjct: 551 KTWVKQVRDVSLCIEDVVDEYEYSKYVAQWIDHFEN 658
>BQ751408 weakly similar to GP|7297956|gb| bun gene product {alt 1}
[Drosophila melanogaster], partial (2%)
Length = 770
Score = 28.1 bits (61), Expect = 8.6
Identities = 15/43 (34%), Positives = 21/43 (47%)
Frame = +3
Query: 123 PKVLAEYMVKNKGFLPSGPSEIHLQRSQIRAIIHSLLPACNSR 165
P L ++ N FL +G +HLQ + + S LP C SR
Sbjct: 513 PTELG*SVIPN*PFLDTGERAVHLQTQHLPS*SRSRLPVCPSR 641
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.141 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,278,191
Number of Sequences: 36976
Number of extensions: 284041
Number of successful extensions: 1372
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 1322
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1368
length of query: 520
length of database: 9,014,727
effective HSP length: 100
effective length of query: 420
effective length of database: 5,317,127
effective search space: 2233193340
effective search space used: 2233193340
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148994.4


