

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148994.1 + phase: 0
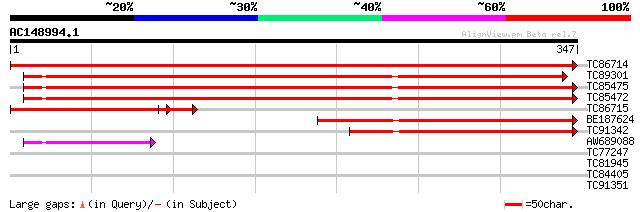
(347 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86714 similar to GP|9759135|dbj|BAB09620.1 amylogenin; reversi... 702 0.0
TC89301 homologue to GP|18077708|emb|CAC83750. reversibly glycos... 374 e-104
TC85475 homologue to PIR|T06507|T06507 reversibly glycosylatable... 366 e-102
TC85472 homologue to GP|2317729|gb|AAC50000.1| reversibly glycos... 362 e-100
TC86715 weakly similar to GP|9759135|dbj|BAB09620.1 amylogenin; ... 185 4e-56
BE187624 similar to GP|6041795|gb| reversibly glycosylated polyp... 165 3e-41
TC91342 similar to PIR|T11577|T11577 type IIIa membrane protein ... 148 3e-36
AW689088 similar to GP|19913103|emb UDP-Glucose:protein transglu... 54 1e-07
TC77247 similar to PIR|T06380|T06380 beta-fructofuranosidase (EC... 29 2.3
TC81945 weakly similar to GP|10177640|dbj|BAB10787. contains sim... 28 5.2
TC84405 similar to PIR|C86353|C86353 protein F2E2.8 [imported] -... 28 6.8
TC91351 homologue to GP|16266960|dbj|BAB70118. NADH dehydrogenas... 28 6.8
>TC86714 similar to GP|9759135|dbj|BAB09620.1 amylogenin; reversibly
glycosylatable polypeptide {Arabidopsis thaliana},
complete
Length = 1954
Score = 702 bits (1812), Expect = 0.0
Identities = 346/347 (99%), Positives = 347/347 (99%)
Frame = +1
Query: 1 MSQVIINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADV 60
MSQVIINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADV
Sbjct: 484 MSQVIINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADV 663
Query: 61 YTNSEIERVVGSSTSIRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQH 120
YTNSEIERVVGSSTSIRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQH
Sbjct: 664 YTNSEIERVVGSSTSIRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQH 843
Query: 121 IVNLKTPATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQAL 180
IVNLKTPATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQAL
Sbjct: 844 IVNLKTPATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQAL 1023
Query: 181 KPTQRNSRYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDI 240
KPTQRNSRYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVE+I
Sbjct: 1024KPTQRNSRYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVENI 1203
Query: 241 WCGLCVKIVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQ 300
WCGLCVKIVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQ
Sbjct: 1204WCGLCVKIVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQ 1383
Query: 301 SATTAEDCVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
SATTAEDCVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA
Sbjct: 1384SATTAEDCVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 1524
>TC89301 homologue to GP|18077708|emb|CAC83750. reversibly glycosylated
polypeptide {Gossypium hirsutum}, partial (93%)
Length = 1270
Score = 374 bits (959), Expect = e-104
Identities = 175/334 (52%), Positives = 241/334 (71%), Gaps = 1/334 (0%)
Frame = +2
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ +W+ F +HLIIV+D + +++PEGF ++Y ++I R
Sbjct: 44 DELDIVIPTIRN--LDFLEQWRQFFQPYHLIIVQDGDPSKVIKVPEGFDYELYNRNDINR 217
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FGF+VSKKKY+ IDDDC AKD +G +DA+ QHI NL TP
Sbjct: 218 ILGPKASCISFKDSACRCFGFIVSKKKYIFTIDDDCFVAKDPSGKEIDALQQHIKNLLTP 397
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TPFFFNTLYDP+R+GADFVRGYPFSLR GV A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 398 STPFFFNTLYDPYREGADFVRGYPFSLREGVPTAISHGLWLNIPDYDAPTQLVKPLERNT 577
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P + P+ G+N+AFNREL+GPAL L+ G+ R+ +D+W G C+K
Sbjct: 578 RYVDAVMTIPKGTLFPMCGMNLAFNRELIGPALYFGLMGEGQPIGRY---DDMWAGWCMK 748
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++ DHL LGVK+GLPY+W ++ N +LKKE++G+ E+++PFFQSV LP+ TT +
Sbjct: 749 VISDHLGLGVKTGLPYIWHSKASNPFVNLKKEYKGIYWQEELIPFFQSVSLPKECTTPQK 928
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLW 341
C IE++K VK +LG VD F K ADAM W+++W
Sbjct: 929 CYIELSKQVKAKLGLVDDYFNKLADAMVTWIEVW 1030
>TC85475 homologue to PIR|T06507|T06507 reversibly glycosylatable
polypeptide 1 - garden pea, partial (95%)
Length = 1726
Score = 366 bits (940), Expect = e-102
Identities = 166/340 (48%), Positives = 244/340 (70%), Gaps = 1/340 (0%)
Frame = +1
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ F ++HLIIV+D + +++P+GF ++Y ++I R
Sbjct: 466 DELDIVIPTIRN--LDFLEMWRPFFEQYHLIIVQDGDPSKVIKVPQGFDYELYNRNDINR 639
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC AKD G+ ++A+ QHI NL +P
Sbjct: 640 ILGPKASCISFKDSACRCFGYMVSKKKYIYTIDDDCFVAKDPTGHEINALEQHIKNLLSP 819
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TPFFFNTLYDP+R GADFVRGYPFSLR GV A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 820 STPFFFNTLYDPYRDGADFVRGYPFSLREGVPTAVSHGLWLNIPDYDAPTQLVKPHERNT 999
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
R+VDAV+T+P + P+ G+N+AF+REL+GPA+ L+ G+ R+ +D+W G C+K
Sbjct: 1000RFVDAVMTIPKGTLFPMCGMNLAFDRELIGPAMYFGLMGDGQPIGRY---DDMWAGWCIK 1170
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHL LGVK+GLPY+W ++ N +LKKE++G+ E+++PFFQ+ L + T+ +
Sbjct: 1171VICDHLGLGVKTGLPYIWHSKASNPFVNLKKEYKGIFWQEEIIPFFQAATLSKDCTSVQK 1350
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
C IE++K VKE+LG +DP F+K ADAM W++ W + ++
Sbjct: 1351CYIELSKQVKEKLGTIDPYFVKLADAMVTWIEAWDEINNS 1470
>TC85472 homologue to GP|2317729|gb|AAC50000.1| reversibly glycosylated
polypeptide-1 {Arabidopsis thaliana}, partial (94%)
Length = 1471
Score = 362 bits (929), Expect = e-100
Identities = 168/340 (49%), Positives = 240/340 (70%), Gaps = 1/340 (0%)
Frame = +3
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ F +HLIIV+D + + +P+GF ++Y ++I R
Sbjct: 66 DELDIVIPTIRN--LDFLEMWRPFFEPYHLIIVQDGDPSKTINVPQGFDYELYNRNDINR 239
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC A D G ++A+ QHI NL P
Sbjct: 240 ILGPKASCISFKDSACRCFGYMVSKKKYIYTIDDDCFVANDPTGKKINALEQHIKNLLCP 419
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TPFFFNTLY+P+R+GADFVRGYPFSLR GV A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 420 STPFFFNTLYEPYREGADFVRGYPFSLREGVPTAVSHGLWLNIPDYDAPTQLVKPLERNT 599
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDA+LT+P + P+ G+N+AF+REL+GPA+ L+ G+ R+ +D+W G C+K
Sbjct: 600 RYVDAILTIPKGTLFPMCGMNLAFDRELIGPAMYFGLMGDGQPIGRY---DDMWAGWCIK 770
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHL LGVK+GLPY++ ++ N +L+KE++G+ ED++PFFQ+V L + ATT +
Sbjct: 771 VICDHLGLGVKTGLPYIYHSKASNPFVNLRKEYKGIFWQEDIIPFFQNVVLSKEATTVQK 950
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
C IE+AK VK++L K+DP F K ADAM W++ W + A
Sbjct: 951 CYIELAKEVKDKLSKIDPYFDKLADAMVTWIESWDELNPA 1070
>TC86715 weakly similar to GP|9759135|dbj|BAB09620.1 amylogenin; reversibly
glycosylatable polypeptide {Arabidopsis thaliana},
partial (32%)
Length = 990
Score = 185 bits (470), Expect(2) = 4e-56
Identities = 93/99 (93%), Positives = 96/99 (96%)
Frame = +1
Query: 1 MSQVIINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADV 60
MSQVIINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADV
Sbjct: 646 MSQVIINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADV 825
Query: 61 YTNSEIERVVGSSTSIRFSGYACRYFGFLVSKKKYVVCI 99
YTNSEIERVVGSSTSIRFSGYACRYFGFLVS++ V C+
Sbjct: 826 YTNSEIERVVGSSTSIRFSGYACRYFGFLVSRR--VCCL 936
Score = 50.8 bits (120), Expect(2) = 4e-56
Identities = 21/24 (87%), Positives = 24/24 (99%)
Frame = +2
Query: 92 KKKYVVCIDDDCVPAKDDAGNVVD 115
+++YVVCIDDDCVPAKDDAGNVVD
Sbjct: 917 QEEYVVCIDDDCVPAKDDAGNVVD 988
>BE187624 similar to GP|6041795|gb| reversibly glycosylated polypeptide-1
{Arabidopsis thaliana}, partial (43%)
Length = 664
Score = 165 bits (417), Expect = 3e-41
Identities = 74/159 (46%), Positives = 112/159 (69%)
Frame = +1
Query: 189 YVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVKI 248
YVDA+LT+P + P+ G+N AF+REL+GPA+ L+ G+ R++ D+W G C+K+
Sbjct: 1 YVDAILTIP*GTLFPMCGMNWAFDRELIGPAMYFGLMGDGQPIGRYD---DMWAGWCIKV 171
Query: 249 VCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAEDC 308
+CDHL LGVK+GLPY++ ++ N +L+KE++G+ ED++PFFQ+V L + ATT + C
Sbjct: 172 ICDHLGLGVKTGLPYIYHSKASNPFVNLRKEYKGIFWQEDIIPFFQNVVLSKEATTVQKC 351
Query: 309 VIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
IE+AK VK++L K+DP F K ADAM W++ W + A
Sbjct: 352 YIELAKEVKDKLSKIDPYFDKLADAMVTWIESWDELNPA 468
>TC91342 similar to PIR|T11577|T11577 type IIIa membrane protein cp-wap13 -
cowpea, partial (41%)
Length = 670
Score = 148 bits (373), Expect = 3e-36
Identities = 65/139 (46%), Positives = 99/139 (70%)
Frame = +1
Query: 209 IAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVKIVCDHLSLGVKSGLPYVWRNE 268
+AF+R+L+GPA+ L+ G+ R++ D+W G C K++CDHL LG+K+GLPY++ ++
Sbjct: 40 LAFDRDLIGPAMYFGLMGDGQPIGRYD---DMWAGWCCKVICDHLGLGIKTGLPYIFHSK 210
Query: 269 RGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAEDCVIEMAKSVKEQLGKVDPMFL 328
N +L+KE++G+ ED++PFFQS+ LP+ ATT + C IEM+K VKE+LGK+DP F
Sbjct: 211 ASNPFVNLRKEYKGIFWQEDIIPFFQSLALPKEATTVQKCYIEMSKQVKEKLGKIDPYFD 390
Query: 329 KAADAMAEWVKLWKSVGSA 347
K ADAM W++ W + A
Sbjct: 391 KLADAMVTWIEAWDQLNPA 447
>AW689088 similar to GP|19913103|emb UDP-Glucose:protein transglucosylase
{Solanum tuberosum}, partial (23%)
Length = 273
Score = 53.5 bits (127), Expect = 1e-07
Identities = 27/82 (32%), Positives = 45/82 (53%), Gaps = 1/82 (1%)
Frame = +2
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DI I + + F+ W+ F +HLIIV+D + +P GF ++Y +I R
Sbjct: 32 DELDIXIPTIRN--LDFLXMWRPXFXPYHLIIVQDGDPSKTXNVPXGFDYELYNRDDINR 205
Query: 69 VVG-SSTSIRFSGYACRYFGFL 89
++G +T I F AC FG++
Sbjct: 206 ILGPKATCISFXDSACPCFGYM 271
>TC77247 similar to PIR|T06380|T06380 beta-fructofuranosidase (EC 3.2.1.26) -
garden pea, partial (96%)
Length = 1943
Score = 29.3 bits (64), Expect = 2.3
Identities = 11/33 (33%), Positives = 18/33 (54%)
Frame = +1
Query: 265 WRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVK 297
W NE + +D +KK W G+ + V+ +S K
Sbjct: 1027 WANESSSVVDDVKKGWSGIHTIPRVIWLHKSGK 1125
>TC81945 weakly similar to GP|10177640|dbj|BAB10787. contains similarity to
heparanase~gene_id:MGG23.2 {Arabidopsis thaliana},
partial (38%)
Length = 1031
Score = 28.1 bits (61), Expect = 5.2
Identities = 14/36 (38%), Positives = 19/36 (51%)
Frame = -1
Query: 12 DIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALK 47
D+ A + N+T WK +FS HL I K P +K
Sbjct: 716 DLPARAFNPNITLTPAFWKKLFSSSHLCIGKHPLVK 609
>TC84405 similar to PIR|C86353|C86353 protein F2E2.8 [imported] -
Arabidopsis thaliana, partial (51%)
Length = 737
Score = 27.7 bits (60), Expect = 6.8
Identities = 11/42 (26%), Positives = 22/42 (52%)
Frame = +1
Query: 29 WKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIERVV 70
WK FS FH +++K +++ +L GF +Y + ++
Sbjct: 547 WKQGFSHFHYVLMKRVSIESKLNGFNGFRGVLYAMRNVSSLL 672
>TC91351 homologue to GP|16266960|dbj|BAB70118. NADH dehydrogenase subunit 1
{Zu cristatus}, partial (4%)
Length = 982
Score = 27.7 bits (60), Expect = 6.8
Identities = 12/41 (29%), Positives = 22/41 (53%)
Frame = -1
Query: 124 LKTPATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSC 164
+KTP PF F+ +Y P F++ P+ ++ D + +C
Sbjct: 814 VKTPKNPFIFSKIYSPKTSLF*FIQVVPYR*KNLSDTSSTC 692
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,023,554
Number of Sequences: 36976
Number of extensions: 153035
Number of successful extensions: 685
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 677
length of query: 347
length of database: 9,014,727
effective HSP length: 97
effective length of query: 250
effective length of database: 5,428,055
effective search space: 1357013750
effective search space used: 1357013750
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148994.1


