

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.7 - phase: 0
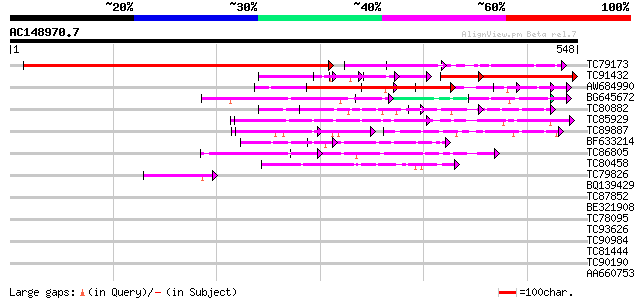
(548 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79173 similar to GP|14532556|gb|AAK64006.1 AT5g57360/MSF19_2 {... 615 e-176
TC91432 homologue to GP|14532556|gb|AAK64006.1 AT5g57360/MSF19_2... 189 1e-70
AW684990 similar to PIR|F96703|F96 unknown protein 35653-33693 ... 236 2e-62
BG645672 similar to GP|7596769|gb| unknown protein {Arabidopsis ... 75 8e-14
TC80882 similar to PIR|G86319|G86319 hypothetical protein AAF984... 69 6e-12
TC85929 similar to GP|6466955|gb|AAF13090.1| unknown protein {Ar... 68 8e-12
TC89887 similar to GP|22136502|gb|AAM91329.1 unknown protein {Ar... 66 4e-11
BF633214 similar to GP|14495224|db contains EST C72594(E1889)~si... 55 9e-08
TC86805 similar to GP|15724206|gb|AAL06496.1 AT5g04420/T32M21_20... 53 3e-07
TC80458 similar to GP|21950739|gb|AAM78582.1 RanGAP1 interacting... 50 2e-06
TC79826 similar to GP|14021816|dbj|BAB48427. hypothetical protei... 42 5e-04
BQ139429 homologue to PIR|E86217|E8 protein T27G7.10 [imported] ... 40 0.003
TC87852 similar to GP|5263329|gb|AAD41431.1| Contains PF|00646 F... 39 0.005
BE321908 similar to GP|22022522|gb AT4g03080/T4I9_4 {Arabidopsis... 37 0.015
TC78095 weakly similar to PIR|G84776|G84776 hypothetical protein... 37 0.026
TC93626 weakly similar to PIR|G84776|G84776 hypothetical protein... 37 0.026
TC90984 similar to GP|18146960|dbj|BAB83170. twin LOV protein 1 ... 34 0.13
TC81444 PIR|C89872|C89872 hypothetical protein [imported] - Stap... 34 0.17
TC90190 similar to GP|3426041|gb|AAC32240.1| unknown protein {Ar... 33 0.22
AA660753 weakly similar to GP|21950739|gb RanGAP1 interacting pr... 30 1.8
>TC79173 similar to GP|14532556|gb|AAK64006.1 AT5g57360/MSF19_2 {Arabidopsis
thaliana}, partial (53%)
Length = 1398
Score = 615 bits (1585), Expect = e-176
Identities = 300/300 (100%), Positives = 300/300 (100%)
Frame = +1
Query: 14 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 73
RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG
Sbjct: 499 RFLQCRGPFAKRRHPLVDSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYG 678
Query: 74 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 133
EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI
Sbjct: 679 EDDEITHVIGIQLFTEANIDLGPLPGSTIKESLKSSGRFHSVLSSLQPPPLGDRNVSRGI 858
Query: 134 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 193
CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL
Sbjct: 859 CGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETTRVL 1038
Query: 194 ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 253
ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN
Sbjct: 1039ETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVN 1218
Query: 254 MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 313
MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF
Sbjct: 1219MQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVF 1398
Score = 63.5 bits (153), Expect = 2e-10
Identities = 33/99 (33%), Positives = 55/99 (55%)
Frame = +1
Query: 324 WREISGLAPPLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVT 383
WR+++ P + S G ++++ GG + ++DTF+LD++ NP W+ + V+
Sbjct: 1114 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVS 1293
Query: 384 WTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVF 422
PP R GHTLS G ++++FGG G L +DVF
Sbjct: 1294 SPPPGRWGHTLSCVNGSRLVVFGGCGTQGLL----NDVF 1398
Score = 54.3 bits (129), Expect = 1e-07
Identities = 46/175 (26%), Positives = 79/175 (44%), Gaps = 1/175 (0%)
Frame = +1
Query: 365 TFLLDMSMENPVWREI-PVTWTPPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFT 423
T L +++ +WR + W S L G K L +G LA+ ++ T
Sbjct: 952 TRLYEVTRNEDLWRMVCQNAWG--SETTRVLETVPGAKRLGWGRLAR---------ELTT 1098
Query: 424 MDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQ 483
++ + WR +T G G+ P R + A ++ G R+++FGG + +
Sbjct: 1099 LEAA----AWRKLTVGG--------GVEPS-RCNFSACAV-GNRVVLFGGEGVNMQPMND 1236
Query: 484 LYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDL 538
++LD P W+ + V PP WGH+ V G+R +V GG G + +L+D+
Sbjct: 1237 TFVLDLNSNNPEWQHVQVSSPPPG-RWGHTLSCVNGSRLVVFGG-CGTQGLLNDV 1395
>TC91432 homologue to GP|14532556|gb|AAK64006.1 AT5g57360/MSF19_2
{Arabidopsis thaliana}, partial (21%)
Length = 755
Score = 189 bits (479), Expect(2) = 1e-70
Identities = 92/95 (96%), Positives = 92/95 (96%)
Frame = +3
Query: 454 PRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHS 513
P L VAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHS
Sbjct: 114 PDLITVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHS 293
Query: 514 TCVVGGTRAIVLGGQTGEEWMLSDLHELSLANSVI 548
TCVVGGTRAIVLGGQTGEEWMLSDLHELSLANSVI
Sbjct: 294 TCVVGGTRAIVLGGQTGEEWMLSDLHELSLANSVI 398
Score = 96.7 bits (239), Expect(2) = 1e-70
Identities = 42/42 (100%), Positives = 42/42 (100%)
Frame = +2
Query: 417 RSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDH 458
RSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDH
Sbjct: 2 RSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDH 127
Score = 54.3 bits (129), Expect = 1e-07
Identities = 26/79 (32%), Positives = 42/79 (52%), Gaps = 2/79 (2%)
Frame = +3
Query: 241 GNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSSPPPG-RWGHTLSCVNGSRLVV 299
G R+++FGG + + ++LD P W+ + V PP WGH+ V G+R +V
Sbjct: 147 GGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIV 326
Query: 300 FGG-CGTQGLLNDVFVLDL 317
GG G + +L+D+ L L
Sbjct: 327 LGGQTGEEWMLSDLHELSL 383
Score = 47.8 bits (112), Expect = 1e-05
Identities = 31/86 (36%), Positives = 48/86 (55%), Gaps = 3/86 (3%)
Frame = +3
Query: 294 GSRLVVFGGCGTQGL--LNDVFVLDLDATPPTWREISGLAPPLPRSW-HSSCTLDGTKLI 350
G R+++FGG GL + +++LD PTWR ++ P +W HS+C + GT+ I
Sbjct: 147 GGRILIFGG-SVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAI 323
Query: 351 VSGGCADSGVLLSDTFLLDMSMENPV 376
V GG +LSD L ++S+ N V
Sbjct: 324 VLGGQTGEEWMLSD--LHELSLANSV 395
Score = 40.8 bits (94), Expect(2) = 2e-06
Identities = 21/66 (31%), Positives = 34/66 (50%), Gaps = 1/66 (1%)
Frame = +3
Query: 343 TLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPS-RLGHTLSVYGGRK 401
+L G ++++ GG S ++LD + E P WR + V PP GH+ V GG +
Sbjct: 138 SLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTR 317
Query: 402 ILMFGG 407
++ GG
Sbjct: 318 AIVLGG 335
Score = 28.9 bits (63), Expect(2) = 2e-06
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 11/44 (25%)
Frame = +2
Query: 310 NDVFVLDLDATPPTWREIS-----------GLAPPLPRSWHSSC 342
+DVF +DL P WR ++ G+APP PR H C
Sbjct: 8 SDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPP-PRLDHXGC 136
>AW684990 similar to PIR|F96703|F96 unknown protein 35653-33693 [imported] -
Arabidopsis thaliana, partial (23%)
Length = 517
Score = 236 bits (601), Expect = 2e-62
Identities = 106/144 (73%), Positives = 126/144 (86%)
Frame = +3
Query: 288 TLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGT 347
TLSC+N S LVVFGGCG QGLLNDVFVLDLDA PTW+E+ G APPLPRSWHSSCT++G+
Sbjct: 3 TLSCLNSSWLVVFGGCGRQGLLNDVFVLDLDAQQPTWKEVFGEAPPLPRSWHSSCTIEGS 182
Query: 348 KLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGHTLSVYGGRKILMFGG 407
KL+VSGGC D+GVLLSDT+LLD++++NP WREIP +WTPPSRLGH+LSVYG KILMFGG
Sbjct: 183 KLVVSGGCTDAGVLLSDTYLLDLTIDNPTWREIPTSWTPPSRLGHSLSVYGRTKILMFGG 362
Query: 408 LAKSGPLRFRSSDVFTMDLSEDEP 431
LAKSG LR RS + +T+DL ++P
Sbjct: 363 LAKSGHLRLRSGEAYTIDLEAEQP 434
Score = 94.0 bits (232), Expect = 1e-19
Identities = 56/143 (39%), Positives = 78/143 (54%), Gaps = 4/143 (2%)
Frame = +3
Query: 237 ACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSR 296
+C + +V+FGG G +ND FVLDL++ P W+ V +PP R H+ + GS+
Sbjct: 9 SCLNSSWLVVFGGCG-RQGLLNDVFVLDLDAQQPTWKEVFGEAPPLPRSWHSSCTIEGSK 185
Query: 297 LVVFGGCGTQG-LLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTKLIVSGGC 355
LVV GGC G LL+D ++LDL PTWREI P R HS TK+++ GG
Sbjct: 186 LVVSGGCTDAGVLLSDTYLLDLTIDNPTWREIPTSWTPPSRLGHSLSVYGRTKILMFGGL 365
Query: 356 ADSGVL---LSDTFLLDMSMENP 375
A SG L + + +D+ E P
Sbjct: 366 AKSGHLRLRSGEAYTIDLEAEQP 434
Score = 75.1 bits (183), Expect = 6e-14
Identities = 51/157 (32%), Positives = 75/157 (47%), Gaps = 3/157 (1%)
Frame = +3
Query: 341 SCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGHTLSVYGGR 400
SC L+ + L+V GGC G LL+D F+LD+ + P W+E+ P R H+ G
Sbjct: 9 SC-LNSSWLVVFGGCGRQG-LLNDVFVLDLDAQQPTWKEVFGEAPPLPRSWHSSCTIEGS 182
Query: 401 KILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPPPRLDHVA 460
K+++ GG +G L SD + +DL+ D P WR + P PP RL H
Sbjct: 183 KLVVSGGCTDAGVLL---SDTYLLDLTIDNPTWREI----------PTSWTPPSRLGHSL 323
Query: 461 VSLPGGRILIFGGSVAGLH---SASQLYILDPTDEKP 494
+IL+FGG H + + Y +D E+P
Sbjct: 324 SVYGRTKILMFGGLAKSGHLRLRSGEAYTIDLEAEQP 434
Score = 70.1 bits (170), Expect = 2e-12
Identities = 45/136 (33%), Positives = 66/136 (48%), Gaps = 1/136 (0%)
Frame = +3
Query: 393 TLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAP 452
TLS +++FGG + G L +DVF +DL +P W+ V G AP
Sbjct: 3 TLSCLNSSWLVVFGGCGRQGLL----NDVFVLDLDAQQPTWKEVFGE-----------AP 137
Query: 453 P-PRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWG 511
P PR H + ++ G ++++ GG S Y+LD T + PTWR + PP G
Sbjct: 138 PLPRSWHSSCTIEGSKLVVSGGCTDAGVLLSDTYLLDLTIDNPTWREIPTSWTPPS-RLG 314
Query: 512 HSTCVVGGTRAIVLGG 527
HS V G T+ ++ GG
Sbjct: 315 HSLSVYGRTKILMFGG 362
Score = 42.0 bits (97), Expect = 6e-04
Identities = 25/77 (32%), Positives = 44/77 (56%), Gaps = 1/77 (1%)
Frame = +3
Query: 468 ILIFGG-SVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLG 526
+++FGG GL + +++LD ++PTW+ + P +W HS+C + G++ +V G
Sbjct: 30 LVVFGGCGRQGL--LNDVFVLDLDAQQPTWKEVFGEAPPLPRSW-HSSCTIEGSKLVVSG 200
Query: 527 GQTGEEWMLSDLHELSL 543
G T +LSD + L L
Sbjct: 201 GCTDAGVLLSDTYLLDL 251
>BG645672 similar to GP|7596769|gb| unknown protein {Arabidopsis thaliana},
partial (32%)
Length = 771
Score = 74.7 bits (182), Expect = 8e-14
Identities = 62/206 (30%), Positives = 86/206 (41%), Gaps = 20/206 (9%)
Frame = +3
Query: 186 GSETTRVLETVPGAKRLGWGRLARELT---------------TLEAAAWRKLTVGGGVEP 230
G E++ L+ G + WG + L+ A W L G
Sbjct: 63 GDESSTTLDPCAGHSLIAWGNKLLSIAGHTKDPSESIQVREFDLQRATWSTLKTYGKPPI 242
Query: 231 SRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQ-VSSPPPGRWGHTL 289
SR S VGN +V+FGG+ +ND +LDL + W + V PP R HT
Sbjct: 243 SRGGQSVSLVGNTLVIFGGQDAKRTLLNDLHILDLETMT--WDEIDAVGVPPSPRSDHTA 416
Query: 290 SCVNGSRLVVFGGCGTQGLLNDVFVLDLD----ATPPTWREISGLAPPLPRSWHSSCTLD 345
+ L++FGG ND+ VLDL + P EI P PR+ H+ T+
Sbjct: 417 AVHVDRYLLIFGGGSHATCYNDLHVLDLQTMEWSRPTQQGEI-----PTPRAGHAGVTVG 581
Query: 346 GTKLIVSGGCADSGVLLSDTFLLDMS 371
IV GG SG S+T +L+MS
Sbjct: 582 ENWFIVGGGDNKSGA--SETVVLNMS 653
Score = 43.9 bits (102), Expect = 2e-04
Identities = 51/196 (26%), Positives = 78/196 (39%), Gaps = 3/196 (1%)
Frame = +3
Query: 335 PRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPP-SRLGHT 393
P + HS L ++G D + + + ++ W + PP SR G +
Sbjct: 90 PCAGHSLIAWGNKLLSIAGHTKDPSESIQ---VREFDLQRATWSTLKTYGKPPISRGGQS 260
Query: 394 LSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPP 453
+S+ G ++ G AK R +D+ +DL + W + G+P P
Sbjct: 261 VSLVGNTLVIFGGQDAK----RTLLNDLHILDL--ETMTWDEIDAVGVP---------PS 395
Query: 454 PRLDHVAVSLPGGRILIFGGSVAGLHSA--SQLYILDPTDEKPTWRILNVPGRPPRFAWG 511
PR DH A +LIFGG G H+ + L++LD + W G P G
Sbjct: 396 PRSDHTAAVHVDRYLLIFGG---GSHATCYNDLHVLDL--QTMEWSRPTQQGEIPTPRAG 560
Query: 512 HSTCVVGGTRAIVLGG 527
H+ VG IV GG
Sbjct: 561 HAGVTVGENWFIVGGG 608
Score = 42.4 bits (98), Expect = 5e-04
Identities = 29/100 (29%), Positives = 45/100 (45%)
Frame = +3
Query: 444 AGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPG 503
AG+ P H ++ G ++L G + Q+ D ++ TW L G
Sbjct: 60 AGDESSTTLDPCAGHSLIAW-GNKLLSIAGHTKDPSESIQVREFDL--QRATWSTLKTYG 230
Query: 504 RPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHELSL 543
+PP G S +VG T ++ GGQ + +L+DLH L L
Sbjct: 231 KPPISRGGQSVSLVGNT-LVIFGGQDAKRTLLNDLHILDL 347
>TC80882 similar to PIR|G86319|G86319 hypothetical protein AAF98413.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1075
Score = 68.6 bits (166), Expect = 6e-12
Identities = 63/188 (33%), Positives = 90/188 (47%), Gaps = 10/188 (5%)
Frame = +3
Query: 281 PPGRWGHTLSCVNGSR-LVVFGGCGTQGL-LNDVFVLDLDATPPTWRE--ISGLAPPLPR 336
P RWGHT + +NG R + VFGG G N V + D TW + I G PP PR
Sbjct: 246 PGKRWGHTCNSINGGRHIYVFGGYGKDNCQTNQVHI--FDTVNQTWSQPAIKG-TPPTPR 416
Query: 337 SWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSM---ENPVWREIPVTWTPPSRLGHT 393
H +C++ G +L V GG D L D +LD S+ ++P+ R P +R GH+
Sbjct: 417 DSH-TCSVVGHRLFVFGG-TDGTNPLKDLHILDTSLQTWDSPIIR----GEGPEAREGHS 578
Query: 394 LSVYGGRKILMFGGLAKSGPLRFRSSDVFTMD---LSEDEPCWRCVTGSGMPGAGNPEGI 450
+V G+++ +FGG KS +++V+ D L+ + W T SG P
Sbjct: 579 AAVV-GKRLYIFGGCGKSAD---NNNEVYYNDLYILNTETLVWMRPTTSGTP-------- 722
Query: 451 APPPRLDH 458
P PR H
Sbjct: 723 -PSPRDSH 743
Score = 61.6 bits (148), Expect = 7e-10
Identities = 56/171 (32%), Positives = 77/171 (44%), Gaps = 9/171 (5%)
Frame = +3
Query: 241 GNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVS-SPPPGRWGHTLSCVNGSRLVV 299
G + +FGG G + N + D + N W + +PP R HT S V G RL V
Sbjct: 288 GRHIYVFGGYGKDNCQTNQVHIFD--TVNQTWSQPAIKGTPPTPRDSHTCSVV-GHRLFV 458
Query: 300 FGGCGTQGLLNDVFVLDLDATPPTWRE--ISGLAPPLPRSWHSSCTLDGTKLIVSGGCAD 357
FGG L D+ + LD + TW I G P R HS+ + G +L + GGC
Sbjct: 459 FGGTDGTNPLKDLHI--LDTSLQTWDSPIIRGEGPE-AREGHSAAVV-GKRLYIFGGCGK 626
Query: 358 SG-----VLLSDTFLLDMSMENPVWREIPVTWTPPS-RLGHTLSVYGGRKI 402
S V +D ++L + E VW + TPPS R H+ S + R I
Sbjct: 627 SADNNNEVYYNDLYIL--NTETLVWMRPTTSGTPPSPRDSHSCSTWKNRII 773
Score = 52.0 bits (123), Expect = 6e-07
Identities = 45/143 (31%), Positives = 63/143 (43%), Gaps = 1/143 (0%)
Frame = +3
Query: 386 PPSRLGHTL-SVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGA 444
P R GHT S+ GGR I +FGG K + T++ + +P +
Sbjct: 246 PGKRWGHTCNSINGGRHIYVFGGYGKDNCQTNQVHIFDTVNQTWSQPAIK---------- 395
Query: 445 GNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGR 504
G P PR H S+ G R+ +FGG+ G + L+ILD + + TW + G
Sbjct: 396 ----GTPPTPRDSHTC-SVVGHRLFVFGGT-DGTNPLKDLHILDTSLQ--TWDSPIIRGE 551
Query: 505 PPRFAWGHSTCVVGGTRAIVLGG 527
P GHS VV G R + GG
Sbjct: 552 GPEAREGHSAAVV-GKRLYIFGG 617
Score = 36.2 bits (82), Expect = 0.033
Identities = 28/82 (34%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Frame = +2
Query: 248 GGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSSPP-PGRWGHTLSCVNGSRLVVFGG-CGT 305
GGE + ++D + D ++ N W+ + S P R GH+ G L VFGG
Sbjct: 782 GGEDGHDYYLSDLHIFDTDTLN--WRELSTSGQLLPPRAGHSTVSF-GKNLFVFGGFTDA 952
Query: 306 QGLLNDVFVLDLDATPPTWREI 327
Q L ND++ LD+D WR I
Sbjct: 953 QNLYNDLYTLDVDT--GIWRNI 1012
Score = 35.8 bits (81), Expect = 0.044
Identities = 17/59 (28%), Positives = 31/59 (51%)
Frame = +2
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQV 277
WR+L+ G + P R S + G + +FGG ND + LD+++ W+++Q+
Sbjct: 848 WRELSTSGQLLPPRAGHSTVSFGKNLFVFGGFTDAQNLYNDLYTLDVDTG--IWRNIQL 1018
>TC85929 similar to GP|6466955|gb|AAF13090.1| unknown protein {Arabidopsis
thaliana}, partial (95%)
Length = 1534
Score = 68.2 bits (165), Expect = 8e-12
Identities = 48/195 (24%), Positives = 83/195 (41%), Gaps = 1/195 (0%)
Frame = +1
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQ 273
L+ AW V G P R + AVG + +FGG +N+ + D +NN W
Sbjct: 301 LDTLAWSVADVSGNTPPPRVGVTMAAVGETIYVFGGRDAEHNELNELYSFDTKTNN--WA 474
Query: 274 HVQVSS-PPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAP 332
+ PP R H+++ + + VFGGCG G LND++ D+ W E+
Sbjct: 475 LISSGDIGPPNRSYHSMT-ADDRNVYVFGGCGVAGRLNDLWAFDV--VDGKWAELPSPGE 645
Query: 333 PLPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTWTPPSRLGH 392
T+ K+ V G A G+ + D +++ + W ++ + P+
Sbjct: 646 SCKGRGGPGLTVAQGKIWVVYGFA--GMEVDDVHFFNLAQK--TWAQVETSGLKPTARSV 813
Query: 393 TLSVYGGRKILMFGG 407
+ G+ I+++GG
Sbjct: 814 FSTCLIGKHIIVYGG 858
Score = 63.9 bits (154), Expect = 1e-10
Identities = 95/349 (27%), Positives = 144/349 (41%), Gaps = 20/349 (5%)
Frame = +1
Query: 218 AWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPM-NDTFVLDLNSNNPEWQHVQ 276
+W KL G ++ +R + + VG +V FGGE P+ N V DL++ W
Sbjct: 157 SWVKLDQRGILQGARSSHAIAVVGQKVYAFGGEFEPRVPVDNKLHVYDLDT--LAWSVAD 330
Query: 277 VS-SPPPGRWGHTLSCVNGSRLVVFGGCGTQ-GLLNDVFVLDLDATPPTWREI-SGLAPP 333
VS + PP R G T++ V G + VFGG + LN+++ D W I SG P
Sbjct: 331 VSGNTPPPRVGVTMAAV-GETIYVFGGRDAEHNELNELY--SFDTKTNNWALISSGDIGP 501
Query: 334 LPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLDMSMENPVWREIPVTW-TPPSRLGH 392
RS+H S T D + V GGC +G L+D + D + + W E+P + R G
Sbjct: 502 PNRSYH-SMTADDRNVYVFGGCGVAG-RLNDLWAFD--VVDGKWAELPSPGESCKGRGGP 669
Query: 393 TLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAP 452
L+V G+ +++G DV +L+ + W V SG+
Sbjct: 670 GLTVAQGKIWVVYGFAG------MEVDDVHFFNLA--QKTWAQVETSGL----------K 795
Query: 453 PPRLDHVAVSLPGGRILIFGGSV---------AGLHSASQLYILDPTDEKPTWRILNVPG 503
P + L G I+++GG + AG S +LY LD E +W L+
Sbjct: 796 PTARSVFSTCLIGKHIIVYGGEIDPSDQGHMGAGQFS-GELYALD--TETLSWTRLDDKV 966
Query: 504 RPPRFAWGHSTCVVGGTR------AIVLGGQTGEEWMLSDLHELSLANS 546
C G +V GG + L D+ L+LA +
Sbjct: 967 DSGDHPGPRGWCAFAGASRGSQEGLLVYGGNSPSNDRLDDIFFLALAQN 1113
Score = 32.3 bits (72), Expect = 0.48
Identities = 31/119 (26%), Positives = 50/119 (41%), Gaps = 15/119 (12%)
Frame = +1
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGE---------GVNMQPMNDTFVLD 264
L W ++ G +R FS C +G ++++GGE G Q + + LD
Sbjct: 751 LAQKTWAQVETSGLKPTARSVFSTCLIGKHIIVYGGEIDPSDQGHMGAG-QFSGELYALD 927
Query: 265 LNSNNPEWQHVQVSS---PPPGRWGHTLSCVNGSR--LVVFGG-CGTQGLLNDVFVLDL 317
+ + +V S P P W GS+ L+V+GG + L+D+F L L
Sbjct: 928 TETLSWTRLDDKVDSGDHPGPRGWCAFAGASRGSQEGLLVYGGNSPSNDRLDDIFFLAL 1104
>TC89887 similar to GP|22136502|gb|AAM91329.1 unknown protein {Arabidopsis
thaliana}, partial (42%)
Length = 1243
Score = 65.9 bits (159), Expect = 4e-11
Identities = 44/146 (30%), Positives = 73/146 (49%), Gaps = 11/146 (7%)
Frame = +2
Query: 219 WRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVL----DLNSNNPEWQH 274
W +L+V + RC +A V R++++GG G M D + L + + P W
Sbjct: 44 WTELSVSATLPQPRCGHTATMVEKRLLVYGGRGGGGPIMGDLWALKGLIEEENETPGWTQ 223
Query: 275 VQVSSPPPG-RWGHTLSCVNGSRLVVFGGCGTQGLLN--DVFVLD---LDATPPTWREIS 328
+++ P R GH+++ G L++FGG GT G L+ D++ D LD TW+ +S
Sbjct: 224 LKLPGQAPSPRCGHSVTNA-GHYLLMFGGHGTGGWLSRYDIYYNDCVVLDRVSATWKRLS 400
Query: 329 -GLAPPLPRSWHSSCTLDGTKLIVSG 353
G PP R++HS + L++ G
Sbjct: 401 VGNEPPPARAYHSMTAIGSRYLLIGG 478
Score = 57.8 bits (138), Expect = 1e-08
Identities = 62/191 (32%), Positives = 94/191 (48%), Gaps = 17/191 (8%)
Frame = +2
Query: 362 LSDTFLLD-MSMENPVWREIPVTWT-PPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSS 419
LSD ++LD +S+E W E+ V+ T P R GHT ++ +++L++GG GP+
Sbjct: 5 LSDVYVLDTISLE---WTELSVSATLPQPRCGHTATMVE-KRLLVYGGRGGGGPIM---G 163
Query: 420 DVFTMD-LSEDE---PCWRCVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGG-S 474
D++ + L E+E P W T +PG AP PR H +V+ G +L+FGG
Sbjct: 164 DLWALKGLIEEENETPGW---TQLKLPGQ------APSPRCGH-SVTNAGHYLLMFGGHG 313
Query: 475 VAGLHSASQLY-----ILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGG-- 527
G S +Y +LD TW+ L+V PP HS + G+R +++GG
Sbjct: 314 TGGWLSRYDIYYNDCVVLDRVSA--TWKRLSVGNEPPPARAYHSMTAI-GSRYLLIGGFD 484
Query: 528 ---QTGEEWML 535
GE W L
Sbjct: 485 GKSTYGEPWWL 517
Score = 44.7 bits (104), Expect = 9e-05
Identities = 30/95 (31%), Positives = 45/95 (46%), Gaps = 7/95 (7%)
Frame = +2
Query: 215 EAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQ------PMNDTFVLDLNSN 268
E W +L + G RC S G+ +++FGG G ND VLD S
Sbjct: 203 ETPGWTQLKLPGQAPSPRCGHSVTNAGHYLLMFGGHGTGGWLSRYDIYYNDCVVLDRVS- 379
Query: 269 NPEWQHVQV-SSPPPGRWGHTLSCVNGSRLVVFGG 302
W+ + V + PPP R H+++ + GSR ++ GG
Sbjct: 380 -ATWKRLSVGNEPPPARAYHSMTAI-GSRYLLIGG 478
>BF633214 similar to GP|14495224|db contains EST C72594(E1889)~similar to
Arabidopsis thaliana chromosome 1 F25I16.5~unknown
protein, partial (16%)
Length = 502
Score = 54.7 bits (130), Expect = 9e-08
Identities = 53/143 (37%), Positives = 69/143 (48%), Gaps = 5/143 (3%)
Frame = +2
Query: 289 LSCVNGSRLVVFGGC--GTQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLD- 345
L V G RLV+FGG G LND+ +LDL W + LP S CTL
Sbjct: 77 LPLVGGERLVIFGGRGEGDANYLNDLHILDLRTM--RWSSFPEVKGDLPVPRDSHCTLAI 250
Query: 346 GTKLIVSGGCADSG-VLLSDTFLLDMSMENPVWREIPVTWTPPS-RLGHTLSVYGGRKIL 403
G KLIV GG DSG D LLD ME W + + P R GH +V G K+
Sbjct: 251 GNKLIVYGG--DSGDQYHGDVSLLD--METMTWTRLKNQGSSPGVRAGHA-AVNVGTKVY 415
Query: 404 MFGGLAKSGPLRFRSSDVFTMDL 426
+ GG+ G R+ +D++ +D+
Sbjct: 416 IIGGV---GDKRY-CNDIWVVDI 472
Score = 43.1 bits (100), Expect = 3e-04
Identities = 27/95 (28%), Positives = 51/95 (53%), Gaps = 1/95 (1%)
Frame = +2
Query: 224 VGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSSPPPG 283
V G + R + A+GN+++++GG+ + Q D +LD+ + W ++ PG
Sbjct: 200 VKGDLPVPRDSHCTLAIGNKLIVYGGDSGD-QYHGDVSLLDMETMT--WTRLKNQGSSPG 370
Query: 284 -RWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDL 317
R GH V G+++ + GG G + ND++V+D+
Sbjct: 371 VRAGHAAVNV-GTKVYIIGGVGDKRYCNDIWVVDI 472
>TC86805 similar to GP|15724206|gb|AAL06496.1 AT5g04420/T32M21_20
{Arabidopsis thaliana}, partial (52%)
Length = 1055
Score = 53.1 bits (126), Expect = 3e-07
Identities = 37/119 (31%), Positives = 55/119 (46%), Gaps = 1/119 (0%)
Frame = +1
Query: 185 WGSETTRVLETVPGAKRLGWGRLARELTTLEAAAWRKLTVGGGVEPSRCNFSACAVGNRV 244
WG + L + G+ R L + +E + + G V +R SA VG+RV
Sbjct: 481 WGEK----LLILGGSSRDTSDTLTVQYIDIETCQFGVIKTSGSVPVARVGQSATLVGSRV 648
Query: 245 VLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSSPPPG-RWGHTLSCVNGSRLVVFGG 302
+LFGGE + +ND VLDL S W ++ S PP R+ H + L++F G
Sbjct: 649 ILFGGEDRRRKLLNDVHVLDLESMT--WDMIKTSQTPPAPRYDHAAAMHGERYLMIFRG 819
Score = 45.4 bits (106), Expect = 5e-05
Identities = 57/216 (26%), Positives = 92/216 (42%), Gaps = 14/216 (6%)
Frame = +1
Query: 272 WQHVQVS-SPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGL 330
W + VS S PP R+ H + V+ +L + GG L+DV V D + TW +
Sbjct: 235 WAPITVSGSRPPARYKHAAAVVD-EKLYIVGGSRNGRHLSDVQVFDFRSL--TWSSLKLK 405
Query: 331 APP-----------LPRSWHSSCTLDGTKLIVSGGCADSGVLLSDTFLLD-MSMENPVWR 378
A LP + + G KL++ GG + SDT + + +E +
Sbjct: 406 ADTGNDNGNSSQENLPATSGHNMIRWGEKLLILGGSSRD---TSDTLTVQYIDIETCQFG 576
Query: 379 EIPVTWTPP-SRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVT 437
I + + P +R+G + ++ G R +++FGG + L +DV +DL + W +
Sbjct: 577 VIKTSGSVPVARVGQSATLVGSR-VILFGGEDRRRKLL---NDVHVLDL--ESMTWDMIK 738
Query: 438 GSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGG 473
S P P PR DH A ++IF G
Sbjct: 739 TSQTP---------PAPRYDHAAAMHGERYLMIFRG 819
Score = 39.3 bits (90), Expect = 0.004
Identities = 34/105 (32%), Positives = 58/105 (54%), Gaps = 4/105 (3%)
Frame = +1
Query: 241 GNRVVLFGGEGVNMQPMNDTFVLD-LNSNNPEWQHVQVS-SPPPGRWGHTLSCVNGSRLV 298
G ++++ GG + +DT + ++ ++ ++ S S P R G + + V GSR++
Sbjct: 484 GEKLLILGGSS---RDTSDTLTVQYIDIETCQFGVIKTSGSVPVARVGQSATLV-GSRVI 651
Query: 299 VFGGCGTQG-LLNDVFVLDLDATPPTWREI-SGLAPPLPRSWHSS 341
+FGG + LLNDV VLDL++ TW I + PP PR H++
Sbjct: 652 LFGGEDRRRKLLNDVHVLDLESM--TWDMIKTSQTPPAPRYDHAA 780
Score = 37.4 bits (85), Expect = 0.015
Identities = 48/182 (26%), Positives = 77/182 (41%), Gaps = 7/182 (3%)
Frame = +1
Query: 369 DMSMENPVWREIPVTWT-PPSRLGHTLSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLS 427
++S +N W I V+ + PP+R H +V K+ + GG L SDV D
Sbjct: 217 ELSYDN--WAPITVSGSRPPARYKHAAAVVD-EKLYIVGGSRNGRHL----SDVQVFD-- 369
Query: 428 EDEPCWRCVTGSGMP---GAGNPEGIAPPPRLDHVA---VSLPGGRILIFGGSVAGLHSA 481
+R +T S + GN G + L + + G ++LI GGS
Sbjct: 370 -----FRSLTWSSLKLKADTGNDNGNSSQENLPATSGHNMIRWGEKLLILGGSSRDTSDT 534
Query: 482 SQLYILDPTDEKPTWRILNVPGRPPRFAWGHSTCVVGGTRAIVLGGQTGEEWMLSDLHEL 541
+ +D E + ++ G P G S +VG +R I+ GG+ +L+D+H L
Sbjct: 535 LTVQYIDI--ETCQFGVIKTSGSVPVARVGQSATLVG-SRVILFGGEDRRRKLLNDVHVL 705
Query: 542 SL 543
L
Sbjct: 706 DL 711
Score = 37.0 bits (84), Expect = 0.020
Identities = 26/83 (31%), Positives = 39/83 (46%)
Frame = +2
Query: 289 LSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSCTLDGTK 348
L C+ L GGC ND+ +LDL + + G PR+ H+ T+D +
Sbjct: 779 LLCMENGTL*FSGGCSHSVFFNDLHLLDLQTMEWSQPQSQGDLVS-PRAGHAGITIDESW 955
Query: 349 LIVSGGCADSGVLLSDTFLLDMS 371
IV GG +G +T +L+MS
Sbjct: 956 FIVGGGDNXNG--CPETLVLNMS 1018
>TC80458 similar to GP|21950739|gb|AAM78582.1 RanGAP1 interacting protein
{Arabidopsis thaliana}, partial (50%)
Length = 1536
Score = 50.1 bits (118), Expect = 2e-06
Identities = 54/198 (27%), Positives = 90/198 (45%), Gaps = 7/198 (3%)
Frame = +1
Query: 244 VVLFGGEGVNMQPMNDTFVLDLNSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGC 303
++LFGGE + +ND + DL S H ++P P R H + + L +FGG
Sbjct: 19 LILFGGEDAKRRKLNDLHMFDLKSLTWLPLHYTGTAPSP-RLNHVAALYDDKVLYIFGGS 195
Query: 304 GTQGLLNDVFVLDLDATPPTWREISGLAPPLPRSWHSSC-TLDGTKLIVSGGCADSGVLL 362
LND++ LD + + ++ G P PR+ C L GTK ++GG
Sbjct: 196 SKSKTLNDLYSLDFETMAWSRVKVRGFHPS-PRA--GCCGVLCGTKWYITGG-GSKKKRH 363
Query: 363 SDTFLLDMSMENPVWREIPVTWTPPSRL----GHTLSV--YGGRKILMFGGLAKSGPLRF 416
+T + D+ W + +T +PPS + G +L + Y + L+ G +K P
Sbjct: 364 GETLIFDIVKNE--W-SVAIT-SPPSSITTNKGFSLVLVQYKEKDYLVAFGGSKKEP--- 522
Query: 417 RSSDVFTMDLSEDEPCWR 434
S+ V ++L ++E R
Sbjct: 523 -SNQVEVLELDKNESALR 573
Score = 34.7 bits (78), Expect = 0.097
Identities = 29/79 (36%), Positives = 38/79 (47%)
Frame = +1
Query: 449 GIAPPPRLDHVAVSLPGGRILIFGGSVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRF 508
G AP PRL+HVA + IFGGS + + + LY LD E W + V G P
Sbjct: 118 GTAPSPRLNHVAALYDDKVLYIFGGS-SKSKTLNDLYSLD--FETMAWSRVKVRGFHPSP 288
Query: 509 AWGHSTCVVGGTRAIVLGG 527
G V+ GT+ + GG
Sbjct: 289 RAG-CCGVLCGTKWYITGG 342
Score = 29.6 bits (65), Expect = 3.1
Identities = 21/104 (20%), Positives = 39/104 (37%)
Frame = +1
Query: 214 LEAAAWRKLTVGGGVEPSRCNFSACAVGNRVVLFGGEGVNMQPMNDTFVLDLNSNNPEWQ 273
L++ W L G R N A ++V+ G + +ND + LD + W
Sbjct: 82 LKSLTWLPLHYTGTAPSPRLNHVAALYDDKVLYIFGGSSKSKTLNDLYSLDFET--MAWS 255
Query: 274 HVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDL 317
V+V P + G++ + GG + + + D+
Sbjct: 256 RVKVRGFHPSPRAGCCGVLCGTKWYITGGGSKKKRHGETLIFDI 387
>TC79826 similar to GP|14021816|dbj|BAB48427. hypothetical protein
{Mesorhizobium loti}, partial (0%)
Length = 728
Score = 42.4 bits (98), Expect = 5e-04
Identities = 21/76 (27%), Positives = 34/76 (44%), Gaps = 4/76 (5%)
Frame = +1
Query: 130 SRGICGIFQLSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAW---- 185
S I + ++L IL L +A+ +S C+ L + + DLW+ +C W
Sbjct: 148 SSSTSSITTMHHDILHTHILTHLDAATLATTASTCSHLRHLCMDNDLWKKICTETWPSLL 327
Query: 186 GSETTRVLETVPGAKR 201
S T+ V+ T P R
Sbjct: 328 DSTTSHVISTFPNGHR 375
>BQ139429 homologue to PIR|E86217|E8 protein T27G7.10 [imported] -
Arabidopsis thaliana, partial (13%)
Length = 531
Score = 39.7 bits (91), Expect = 0.003
Identities = 39/154 (25%), Positives = 65/154 (41%), Gaps = 1/154 (0%)
Frame = +1
Query: 394 LSVYGGRKILMFGGLAKSGPLRFRSSDVFTMDLSEDEPCWRCVTGSGMPGAGNPEGIAPP 453
+++ G R ++ GG PL +DV+ +D + WR + PEG PP
Sbjct: 1 MALVGQRYLMAIGGNDGKRPL----ADVWALDTAAKPYEWRKL---------EPEGEGPP 141
Query: 454 PRLDHVAVSLPGGRILIFGG-SVAGLHSASQLYILDPTDEKPTWRILNVPGRPPRFAWGH 512
P + A + G +L+ GG + + AS + D + W I PG P + H
Sbjct: 142 PCMYATASARSDGLLLLCGGRDASSVPLASAYGLAKHRDGRWEWAI--APGVSPSSRYQH 315
Query: 513 STCVVGGTRAIVLGGQTGEEWMLSDLHELSLANS 546
+ V R V GG G M+ D +++ ++
Sbjct: 316 AAVFV-NARLHVSGGALGGGRMVEDSSSVAVLDT 414
>TC87852 similar to GP|5263329|gb|AAD41431.1| Contains PF|00646 F-box
domain. ESTs gb|Z37267 gb|R90412 gb|Z37268 and
gb|T88189 come from this, partial (97%)
Length = 1470
Score = 38.9 bits (89), Expect = 0.005
Identities = 18/47 (38%), Positives = 24/47 (50%)
Frame = +2
Query: 139 LSDEVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAW 185
L DE+L ++ AR+TP D+ S VC + RN WR C W
Sbjct: 359 LPDELL-FEVFARMTPYDMGKASCVCRKWRYTIRNPAFWRNACLKGW 496
>BE321908 similar to GP|22022522|gb AT4g03080/T4I9_4 {Arabidopsis thaliana},
partial (12%)
Length = 325
Score = 37.4 bits (85), Expect = 0.015
Identities = 34/120 (28%), Positives = 54/120 (44%), Gaps = 15/120 (12%)
Frame = +3
Query: 389 RLGHTLSVYG-----GRKILMFGGL------AKSGP---LRFRSSDVFTMDLSEDEPCWR 434
R GHTL+ G ++++FGG + S P L ++ V D+ D W
Sbjct: 3 RCGHTLTAVAATKSHGPRLILFGGATAIEGGSTSAPGIRLAGVTNSVHAYDV--DSKKWT 176
Query: 435 CVTGSGMPGAGNPEGIAPPPRLDHVAVSLPGGRILIFGGSVAGL-HSASQLYILDPTDEK 493
+ +G P P PR H A ++ G +++F G + HS LY+LD T++K
Sbjct: 177 RIKPAGDP---------PSPRAAHAAATV--GTMVVFQGGIGPAGHSTDDLYVLDLTNDK 323
>TC78095 weakly similar to PIR|G84776|G84776 hypothetical protein At2g36090
[imported] - Arabidopsis thaliana, partial (19%)
Length = 1467
Score = 36.6 bits (83), Expect = 0.026
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Frame = +2
Query: 143 VLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGS----ETTRVLETVPG 198
+L IL+RL + SVSS T L+ + LW+ + + W S T ++ T P
Sbjct: 281 ILQSHILSRLDGTSLTSVSSTATHLHNLCTEHHLWQKIITSTWPSLNNPSATSLISTFPS 460
Query: 199 AKR 201
+ R
Sbjct: 461 SHR 469
>TC93626 weakly similar to PIR|G84776|G84776 hypothetical protein At2g36090
[imported] - Arabidopsis thaliana, partial (25%)
Length = 805
Score = 36.6 bits (83), Expect = 0.026
Identities = 14/49 (28%), Positives = 27/49 (54%)
Frame = +3
Query: 142 EVLSLKILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGSETT 190
+++ IL L +AS +S C++L ++ ++ LW +C + W S T
Sbjct: 66 DIIQTHILTYLDGSSLASAASTCSQLNTLSSHDHLWTNICHSTWPSTNT 212
>TC90984 similar to GP|18146960|dbj|BAB83170. twin LOV protein 1
{Arabidopsis thaliana}, partial (29%)
Length = 737
Score = 34.3 bits (77), Expect = 0.13
Identities = 16/56 (28%), Positives = 29/56 (51%)
Frame = +2
Query: 31 DSSVISEIRKCIDEGVEFQGELLNFRKDGSPLMNRLRLTPIYGEDDEITHVIGIQL 86
D S + IR+ I +LN+RKD S N L ++P+ ++ + +G+Q+
Sbjct: 185 DDSALQLIRESIKTEKLCTVRILNYRKDKSSFWNLLHISPVRDATGKVAYFVGVQI 352
>TC81444 PIR|C89872|C89872 hypothetical protein [imported] - Staphylococcus
aureus (strain N315), partial (16%)
Length = 1434
Score = 33.9 bits (76), Expect = 0.17
Identities = 14/40 (35%), Positives = 21/40 (52%)
Frame = -2
Query: 148 ILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWGS 187
IL RL +AS++ C +++ E LW VC + W S
Sbjct: 1133 ILRRLDGPALASLACTCAAFCSISKEESLWENVCASVWPS 1014
>TC90190 similar to GP|3426041|gb|AAC32240.1| unknown protein {Arabidopsis
thaliana}, partial (60%)
Length = 1626
Score = 33.5 bits (75), Expect = 0.22
Identities = 14/39 (35%), Positives = 23/39 (58%)
Frame = +2
Query: 148 ILARLTPRDIASVSSVCTRLYEVTRNEDLWRMVCQNAWG 186
IL +L+P ++ +V+ VC L E R++ LW + WG
Sbjct: 575 ILEKLSPSELCNVAQVCKSLREKCRSDYLWEKHMKMKWG 691
>AA660753 weakly similar to GP|21950739|gb RanGAP1 interacting protein
{Arabidopsis thaliana}, partial (14%)
Length = 501
Score = 30.4 bits (67), Expect = 1.8
Identities = 15/63 (23%), Positives = 30/63 (46%)
Frame = +3
Query: 266 NSNNPEWQHVQVSSPPPGRWGHTLSCVNGSRLVVFGGCGTQGLLNDVFVLDLDATPPTWR 325
+ ++ W + ++ P + +CV ++++V GG GL V VL+ D +W
Sbjct: 294 SGSSXNWMVLSIAGDKPTPXSYHAACVXENKMIVVGGESGNGLXXXVXVLNFDTF--SWT 467
Query: 326 EIS 328
+S
Sbjct: 468 TVS 476
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,238,022
Number of Sequences: 36976
Number of extensions: 316630
Number of successful extensions: 1630
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 1547
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1601
length of query: 548
length of database: 9,014,727
effective HSP length: 101
effective length of query: 447
effective length of database: 5,280,151
effective search space: 2360227497
effective search space used: 2360227497
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148970.7


