

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.5 + phase: 0
(402 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
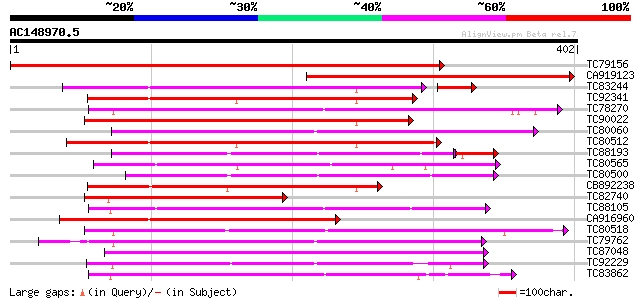
Score E
Sequences producing significant alignments: (bits) Value
TC79156 weakly similar to GP|11034603|dbj|BAB17127. putative rec... 632 0.0
CA919123 weakly similar to PIR|T01537|T015 S-receptor kinase (EC... 278 3e-75
TC83244 weakly similar to GP|11034602|dbj|BAB17126. putative rec... 199 9e-56
TC92341 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Or... 211 3e-55
TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Ar... 211 5e-55
TC90022 similar to GP|14587271|dbj|BAB61189. putative receptor-l... 206 1e-53
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 201 3e-52
TC80512 weakly similar to PIR|A96693|A96693 probable receptor se... 201 4e-52
TC88193 similar to PIR|G96602|G96602 probable receptor protein k... 186 1e-51
TC80565 weakly similar to GP|14335072|gb|AAK59800.1 AT5g20050/F2... 199 1e-51
TC80500 similar to PIR|G96602|G96602 probable receptor protein k... 199 1e-51
CB892238 weakly similar to GP|21741394|em OSJNBb0051N19.7 {Oryza... 198 3e-51
TC82740 weakly similar to PIR|F86420|F86420 hypothetical protein... 198 4e-51
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 184 4e-47
CA916960 weakly similar to GP|14090203|db putative receptor kina... 184 5e-47
TC80518 weakly similar to PIR|T49986|T49986 lectin-like protein ... 179 1e-45
TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.... 179 1e-45
TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsi... 179 2e-45
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 179 2e-45
TC83862 similar to PIR|T05270|T05270 probable serine/threonine-s... 176 1e-44
>TC79156 weakly similar to GP|11034603|dbj|BAB17127. putative receptor
kinase {Oryza sativa (japonica cultivar-group)}, partial
(18%)
Length = 1246
Score = 632 bits (1629), Expect = 0.0
Identities = 308/308 (100%), Positives = 308/308 (100%)
Frame = +1
Query: 1 MFSYTQIPTTEKDNQISSNSFSSTNNIEVVVQTDSKHEFATMERIFSNINREKPVRFTPE 60
MFSYTQIPTTEKDNQISSNSFSSTNNIEVVVQTDSKHEFATMERIFSNINREKPVRFTPE
Sbjct: 322 MFSYTQIPTTEKDNQISSNSFSSTNNIEVVVQTDSKHEFATMERIFSNINREKPVRFTPE 501
Query: 61 KLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYH 120
KLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYH
Sbjct: 502 KLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYH 681
Query: 121 INLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYL 180
INLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYL
Sbjct: 682 INLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYL 861
Query: 181 HEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEM 240
HEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEM
Sbjct: 862 HEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEM 1041
Query: 241 WKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALC 300
WKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALC
Sbjct: 1042WKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALC 1221
Query: 301 EIEEKDSE 308
EIEEKDSE
Sbjct: 1222EIEEKDSE 1245
>CA919123 weakly similar to PIR|T01537|T015 S-receptor kinase (EC 2.7.1.-)
homolog 1 precursor - Arabidopsis thaliana, partial (8%)
Length = 793
Score = 278 bits (710), Expect = 3e-75
Identities = 137/191 (71%), Positives = 153/191 (79%), Gaps = 1/191 (0%)
Frame = -3
Query: 211 FGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHF- 269
+ ++KLRSRES+I +N F+G YAAPEMWK YP TYKCDVYSFGI+LFEIVGRR HF
Sbjct: 779 YWISKLRSRESHIAMNNRFKGKAAYAAPEMWKAYPETYKCDVYSFGIVLFEIVGRRTHFI 600
Query: 270 DSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRP 329
D +YSESQ WF + TWEMFEN+ELV ML C IEEKD+E AERMLKVA WCVQYSP+DRP
Sbjct: 599 DPTYSESQIWFSKRTWEMFENSELVAMLEFCGIEEKDTEKAERMLKVAQWCVQYSPDDRP 420
Query: 330 LMSTVVKMLEGEIDISSPPFPFHNLVPAKENSTQEGSTADSDTTTSSWRTESSRESGFKT 389
LMSTVVKMLEGE +I PFPFHN V KENSTQEGSTA S+T TSSW T S RESGF+
Sbjct: 419 LMSTVVKMLEGEKEILQAPFPFHNRVSVKENSTQEGSTAYSNTATSSWHT*SLRESGFRA 240
Query: 390 KHNAFEIETAT 400
K N F+IE T
Sbjct: 239 KQNTFQIEKPT 207
>TC83244 weakly similar to GP|11034602|dbj|BAB17126. putative receptor
kinase {Oryza sativa (japonica cultivar-group)}, partial
(36%)
Length = 1366
Score = 199 bits (506), Expect(2) = 9e-56
Identities = 115/262 (43%), Positives = 156/262 (58%), Gaps = 4/262 (1%)
Frame = +1
Query: 38 EFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVL 97
+ A +E+ + KP RFT + IT + LG GA G VFKG LS VAVKVL
Sbjct: 484 DIARIEKFLEDYRALKPTRFTYADIKRITNGFKESLGEGAHGSVFKGMLSQEILVAVKVL 663
Query: 98 NCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKN 157
N G F EV T+G+ +H+N+V+L GFC RALVY++ NGSL ++ +N
Sbjct: 664 N-ETQGDGNDFINEVGTMGKIHHVNVVRLLGFCADGFHRALVYDFFPNGSLQNFLAPPEN 840
Query: 158 RNDF-DFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKL 216
+ F +KL +IA+G A+GI YLH C HRI+H+DI P NVL+D L PKI DFGLAKL
Sbjct: 841 KEVFLGREKLQRIALGVARGIEYLHIGCDHRILHFDINPHNVLIDDNLSPKITDFGLAKL 1020
Query: 217 RSRESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSYS 274
+ + T RGT GY APE++ V+YK D+YS+G+LL E+VG R++ + S
Sbjct: 1021CPKNQSTVSMTAARGTLGYIAPEVFSRNFGNVSYKSDIYSYGMLLLEMVGGRKNTNQSAK 1200
Query: 275 ES-QQWFPRWTWEMFENNELVV 295
E+ Q +P W + E E+ V
Sbjct: 1201ETFQVLYPEWIHNLIEG*EVRV 1266
Score = 35.8 bits (81), Expect(2) = 9e-56
Identities = 14/28 (50%), Positives = 20/28 (71%)
Frame = +2
Query: 304 EKDSEIAERMLKVALWCVQYSPNDRPLM 331
E D IA+++ V LWC+Q++P DRP M
Sbjct: 1280 EGDVRIAKKLALVGLWCIQWNPVDRPSM 1363
>TC92341 similar to GP|21741394|emb|CAD39676. OSJNBb0051N19.7 {Oryza
sativa}, partial (60%)
Length = 779
Score = 211 bits (538), Expect = 3e-55
Identities = 112/239 (46%), Positives = 158/239 (65%), Gaps = 5/239 (2%)
Frame = +2
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITI 115
R+ ++ +T LG G FGVV+KG+L NG VAVK+LN + G E+F EV +I
Sbjct: 29 RYKYSEIKNMTNSLKDKLGQGGFGVVYKGKLFNGCLVAVKILN-VSKGNGEEFINEVASI 205
Query: 116 GRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRN--DFDFQKLHKIAIGT 173
RT H+N+V L GFCF +K+ALVYE++ NGSLDK+I+ + + KL+KIA G
Sbjct: 206 SRTSHVNVVTLLGFCFEGNKKALVYEFMSNGSLDKFIYNKELETIASLSWDKLYKIAKGI 385
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
A+G+ YLH C RI+H+DIKP N+LLD L PKI+DFGLAKL R+ +I + RGT
Sbjct: 386 ARGLEYLHGGCTTRILHFDIKPHNILLDDNLCPKISDFGLAKLCLRKESIVSMSDQRGTM 565
Query: 234 GYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSYSE-SQQWFPRWTWEMFE 289
GY APE+W + V++K DVYS+G++L E+VG R++ ++ S S+ +FP W ++ E
Sbjct: 566 GYVAPEVWNRHFGGVSHKSDVYSYGMILLEMVGGRKNINADASRTSEIYFPHWVYKRLE 742
>TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1463
Score = 211 bits (536), Expect = 5e-55
Identities = 135/358 (37%), Positives = 194/358 (53%), Gaps = 22/358 (6%)
Frame = +3
Query: 57 FTPEKLDEITEKYSTI--LGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVIT 114
+T ++L ++ +S +G G FG V+KG L G+ A+KVL+ ++F E+
Sbjct: 273 YTYKELKIASDNFSPANKIGEGGFGSVYKGVLKGGKLAAIKVLSTESKQGVKEFLTEINV 452
Query: 115 IGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRND-FDFQKLHKIAIGT 173
I H NLV LYG C D R LVY Y+EN SL + + + N FD+Q +I +G
Sbjct: 453 ISEIKHENLVILYGCCVEGDHRILVYNYLENNSLSQTLLAGGHSNIYFDWQTRRRICLGV 632
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
A+G+A+LHEE I+H DIK N+LLD L PKI+DFGLAKL ++T GT
Sbjct: 633 ARGLAFLHEEVLPHIVHRDIKASNILLDKDLTPKISDFGLAKLIPSYMT-HVSTRVAGTI 809
Query: 234 GYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNEL 293
GY APE +T K D+YSFG+LL EIV R + ++ + Q+ TW+++E EL
Sbjct: 810 GYLAPEYAIRGQLTRKADIYSFGVLLVEIVSGRSNTNTRLPIADQYILETTWQLYERKEL 989
Query: 294 VVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPFHN 353
++ + E D+E A ++LK+AL C Q +P RP MS+VVKML GE+DI+
Sbjct: 990 AQLVDISLNGEFDAEEACKILKIALLCTQDTPKLRPTMSSVVKMLTGEMDINETKITKPG 1169
Query: 354 LV----------PAKE-------NSTQEGSTADSD--TTTSSWRTESSRESGFKTKHN 392
L+ P K +S S++DS TT S + S+ S F K++
Sbjct: 1170LISDVMDLKIREPKKNINMGTAPSSYNASSSSDSQGTTTISLAASSSATTSSFTVKYD 1343
>TC90022 similar to GP|14587271|dbj|BAB61189. putative receptor-like kinase
{Oryza sativa (japonica cultivar-group)}, partial (31%)
Length = 773
Score = 206 bits (524), Expect = 1e-53
Identities = 105/237 (44%), Positives = 158/237 (66%), Gaps = 4/237 (1%)
Frame = +1
Query: 54 PVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNC-LDMGMEEQFKAEV 112
P+R++ ++ ++T + LG G +G V+KG+L +G +VA+K+L G + F EV
Sbjct: 7 PIRYSYTEIKKMTNGFKDKLGEGGYGKVYKGKLRSGPSVAIKMLGKHKGKGNGQDFINEV 186
Query: 113 ITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIG 172
TIGR +H N+V+L GFC KRALVY+++ NGSLDKYI ++ ++++++I++
Sbjct: 187 ATIGRIHHTNVVRLIGFCVEGSKRALVYDFMPNGSLDKYISSREDHISLTYKQIYEISLA 366
Query: 173 TAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGT 232
A+GIAYLH+ C +I+H+DIKP N+LLD K++DFGLAKL E++I T RGT
Sbjct: 367 VARGIAYLHQGCDMQILHFDIKPHNILLDQDFIAKVSDFGLAKLYPIENSIVTMTAARGT 546
Query: 233 RGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQ-WFPRWTWE 286
GY APE++ V+YK DVYSFG+LL EI RRR+ +S+ +S Q +FP W ++
Sbjct: 547 IGYMAPELFYKNIGKVSYKADVYSFGMLLMEIANRRRNLNSNADDSSQIFFPYWIYQ 717
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 201 bits (512), Expect = 3e-52
Identities = 123/304 (40%), Positives = 168/304 (54%), Gaps = 1/304 (0%)
Frame = +1
Query: 73 LGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFH 132
+G G FG V+KG LS+G +AVK L+ +F E+ I H NLVKLYG C
Sbjct: 211 IGEGGFGPVYKGVLSDGAVIAVKQLSSKSKQGNREFVNEIGMISALQHPNLVKLYGCCIE 390
Query: 133 RDKRALVYEYVENGSLDKYIFGS-KNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHY 191
++ LVYEY+EN SL + +FG + R + D++ KI +G A+G+AYLHEE + +I+H
Sbjct: 391 GNQLLLVYEYMENNSLARALFGKPEQRLNLDWRTRMKICVGIARGLAYLHEESRLKIVHR 570
Query: 192 DIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCD 251
DIK NVLLD L KI+DFGLAKL E N ++T GT GY APE +T K D
Sbjct: 571 DIKATNVLLDKNLNAKISDFGLAKL-DEEENTHISTRIAGTIGYMAPEYAMRGYLTDKAD 747
Query: 252 VYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAE 311
VYSFG++ EIV + + E + W + + E L+ ++ + SE A
Sbjct: 748 VYSFGVVALEIVSGMSNTNYRPKEEFVYLLDWAYVLQEQGNLLELVDPTLGSKYSSEEAM 927
Query: 312 RMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPFHNLVPAKENSTQEGSTADSD 371
RML++AL C SP RP MS+VV MLEG I +P + E + DS
Sbjct: 928 RMLQLALLCTNPSPTLRPPMSSVVSMLEGNTPIQAPIIKRSDSTAGARFKAFELLSQDSQ 1107
Query: 372 TTTS 375
TT++
Sbjct: 1108TTST 1119
>TC80512 weakly similar to PIR|A96693|A96693 probable receptor
serine/threonine kinase PR5K T4O24.7 [imported] -
Arabidopsis thaliana, partial (35%)
Length = 1077
Score = 201 bits (511), Expect = 4e-52
Identities = 115/274 (41%), Positives = 171/274 (61%), Gaps = 8/274 (2%)
Frame = +1
Query: 41 TMERIF-SNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNC 99
TM +F + N R++ ++ IT + LG G +GVV+K L++G VAVKV+N
Sbjct: 247 TMVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVIN- 423
Query: 100 LDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRN 159
G E+F EV +I RT H+N+V L G+C+ +KRAL+YE++ GSL+K+I+ S +
Sbjct: 424 ESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLNKFIYKSGFPD 603
Query: 160 ---DFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKL 216
DFD L +IAIG A+G+ YLH+ C RI+H DIKP+N+LLD PKI+DFGLAK+
Sbjct: 604 AVCDFDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKI 783
Query: 217 RSRESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSYS 274
+I RGT GY APE++ V+YK DVYS+G+L+ E++G R+++ + S
Sbjct: 784 CQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGS 963
Query: 275 -ESQQWFPRWTW-EMFENNELVVMLALCEIEEKD 306
S+ +FP W + ++ + N+L L + E EE D
Sbjct: 964 CTSEMYFPDWIYKDLEQGNDLXNSLTISE-EEND 1062
>TC88193 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(20%)
Length = 1815
Score = 186 bits (471), Expect(2) = 1e-51
Identities = 103/246 (41%), Positives = 147/246 (58%)
Frame = +3
Query: 73 LGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFH 132
LG G FG V+KG L++G VAVK L+ + QF AE+ TI H NLVKLYG C
Sbjct: 594 LGEGGFGPVYKGTLNDGRFVAVKQLSIGSHQGKSQFIAEIATISAVQHRNLVKLYGCCIE 773
Query: 133 RDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYD 192
+KR LVYEY+EN SLD+ +FG N ++ + + +G A+G+ YLHEE + RI+H D
Sbjct: 774 GNKRLLVYEYLENKSLDQALFG--NVLFLNWSTRYDVCMGVARGLTYLHEESRLRIVHRD 947
Query: 193 IKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDV 252
+K N+LLD +L PK++DFGLAKL + ++T GT GY APE +T K DV
Sbjct: 948 VKASNILLDSELVPKLSDFGLAKLYD-DKKTHISTRVAGTIGYLAPEYAMRGRLTEKADV 1124
Query: 253 YSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAER 312
+SFG++ E+V R + DSS E + + W W++ E N + L + E + E ER
Sbjct: 1125FSFGVVALELVSGRPNSDSSLEEDKMYLLDWAWQLHERN-CINDLIDPRLSEFNMEEVER 1301
Query: 313 MLKVAL 318
++ + +
Sbjct: 1302LVGIGI 1319
Score = 35.8 bits (81), Expect(2) = 1e-51
Identities = 19/35 (54%), Positives = 23/35 (65%), Gaps = 5/35 (14%)
Frame = +1
Query: 317 ALW-----CVQYSPNDRPLMSTVVKMLEGEIDISS 346
ALW C Q SPN RP MS VV ML G+I++S+
Sbjct: 1300 ALWE*GLLCTQTSPNLRPSMSRVVAMLLGDIEVST 1404
>TC80565 weakly similar to GP|14335072|gb|AAK59800.1 AT5g20050/F28I16_200
{Arabidopsis thaliana}, partial (81%)
Length = 1581
Score = 199 bits (507), Expect = 1e-51
Identities = 113/303 (37%), Positives = 174/303 (57%), Gaps = 14/303 (4%)
Frame = +2
Query: 60 EKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTY 119
+ L + + + I+G G+ VFKG L++G +VAVK ++ + G E +F++EV I
Sbjct: 401 KNLKKQXDNFQAIIGKGSSASVFKGILNDGTSVAVKRIHGEERG-EREFRSEVSAIASVQ 577
Query: 120 HINLVKLYGFCFH-RDKRALVYEYVENGSLDKYIFGSKNRND-----FDFQKLHKIAIGT 173
H+NLV+L+G+C R LVYE++ NGSLD +IF K + +K+AI
Sbjct: 578 HVNLVRLFGYCNSPTPPRYLVYEFIPNGSLDCWIFPVKETRTRRCGCLPWNLRYKVAIDV 757
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
AK ++YLH +C+ ++H D+KPEN+LLD + ++DFGL+KL ++ + L T RGTR
Sbjct: 758 AKALSYLHHDCRSTVLHLDVKPENILLDENYKALVSDFGLSKLVGKDESQVLTT-IRGTR 934
Query: 234 GYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFD------SSYSESQQWFPRWTWEM 287
GY APE ++ K D+YSFG++L EIVG RR+ + + Q+FP+ E
Sbjct: 935 GYLAPEWLLERGISEKTDIYSFGMVLLEIVGGRRNVSKVEDPRDNTKKKWQFFPKIVNEK 1114
Query: 288 FENNEL--VVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDIS 345
+L +V + + D +R++ +ALWC+Q P RP M VV MLEG + +
Sbjct: 1115LREGKLMEIVDQRVVDFGGVDENEVKRLVFIALWCIQEKPRLRPSMVEVVDMLEGRVRVE 1294
Query: 346 SPP 348
PP
Sbjct: 1295EPP 1303
>TC80500 similar to PIR|G96602|G96602 probable receptor protein kinase
F14G9.24 [imported] - Arabidopsis thaliana, partial
(13%)
Length = 1015
Score = 199 bits (507), Expect = 1e-51
Identities = 110/264 (41%), Positives = 159/264 (59%)
Frame = +1
Query: 83 KGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEY 142
+G L++G +VAVK L+ + QF AE+ TI H NLVKLYG C KR LVYEY
Sbjct: 19 QGILNDGRDVAVKQLSIGSHQGKSQFVAEIATISAVQHRNLVKLYGCCIEGSKRLLVYEY 198
Query: 143 VENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDM 202
+EN SLD+ +FG N ++ + I +G A+G+ YLHEE + RI+H D+K N+LLD
Sbjct: 199 LENKSLDQALFG--NVLFLNWSTRYDICMGVARGLTYLHEESRLRIVHRDVKASNILLDS 372
Query: 203 KLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEI 262
+L PKI+DFGLAKL + ++T GT GY APE +T K DV+SFG++ E+
Sbjct: 373 ELVPKISDFGLAKLYD-DKKTHISTRVAGTIGYLAPEYAMRGHLTEKADVFSFGVVALEL 549
Query: 263 VGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQ 322
V R + DS+ + + W W++ E N + ++ + E + E +R++ +AL C Q
Sbjct: 550 VSGRPNSDSTLEGEKMYLLEWAWQLHERNTINELID-PRLSEFNKEEVQRLVGIALLCTQ 726
Query: 323 YSPNDRPLMSTVVKMLEGEIDISS 346
SP RP MS VV ML G+I++ +
Sbjct: 727 TSPTLRPSMSRVVAMLSGDIEVGT 798
>CB892238 weakly similar to GP|21741394|em OSJNBb0051N19.7 {Oryza sativa},
partial (61%)
Length = 726
Score = 198 bits (504), Expect = 3e-51
Identities = 101/213 (47%), Positives = 146/213 (68%), Gaps = 4/213 (1%)
Frame = +3
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITI 115
R+ ++ ++T+ + LG G FGVV+KG+L NG +VA+K+LN G E+F EV +I
Sbjct: 87 RYKYSEIKKMTDSFKVKLGQGGFGVVYKGKLFNGCHVAIKILNS-SKGNGEEFINEVSSI 263
Query: 116 GRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIF--GSKNRNDFDFQKLHKIAIGT 173
RT H+N+V L GFCF K+AL+YE++ NGSLDK+I+ G + ++ L++IA G
Sbjct: 264 TRTSHVNVVTLLGFCFEGTKKALIYEFMSNGSLDKFIYNKGPETIASLSWENLYQIAKGI 443
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
A+G+ YLH C RI+H+DIKP N+LLD L PKI+DFGLAKL ++ +I + RGT
Sbjct: 444 ARGLEYLHRGCTTRILHFDIKPHNILLDENLCPKISDFGLAKLCPKQESIISMSDQRGTM 623
Query: 234 GYAAPEMWKPY--PVTYKCDVYSFGILLFEIVG 264
GY APE+W + V++K DVYS+G++L E+VG
Sbjct: 624 GYVAPEVWNRHFGGVSHKSDVYSYGMMLLEMVG 722
>TC82740 weakly similar to PIR|F86420|F86420 hypothetical protein AAG12910.1
[imported] - Arabidopsis thaliana, partial (5%)
Length = 615
Score = 198 bits (503), Expect = 4e-51
Identities = 98/157 (62%), Positives = 116/157 (73%), Gaps = 13/157 (8%)
Frame = +1
Query: 54 PVRFTPEKLDEITEK-------------YSTILGSGAFGVVFKGELSNGENVAVKVLNCL 100
P+RFTPE+LDEIT K + T+LGS A G FKGELSNG+ VAVK NC
Sbjct: 145 PMRFTPEQLDEITNKCNYTSFSFGKMRQHKTMLGSSASGTDFKGELSNGQKVAVKDFNCF 324
Query: 101 DMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRND 160
MGMEEQF +V TI R +H N++K+YGFC HRDK ALVYEYVENGSL KY+F SKN +D
Sbjct: 325 IMGMEEQFIEDVSTIARIHHTNIIKVYGFCLHRDKNALVYEYVENGSLAKYLFNSKNCDD 504
Query: 161 FDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPEN 197
+F+KL IAIG AKG +YLHE C++RIIHYDI+PEN
Sbjct: 505 LNFEKLRDIAIGIAKGNSYLHEGCEYRIIHYDIRPEN 615
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 184 bits (468), Expect = 4e-47
Identities = 117/290 (40%), Positives = 163/290 (55%), Gaps = 5/290 (1%)
Frame = +2
Query: 57 FTPEKLDEITEKYS--TILGSGAFGVVFKGELSNGENVAVK-VLNCLDMGMEEQFKAEVI 113
FT L+ T K+S I+G G +GVV++G+L NG VA+K +LN L E++F+ EV
Sbjct: 689 FTLRDLELATNKFSKDNIIGEGGYGVVYQGQLINGNPVAIKKLLNNLGQA-EKEFRVEVE 865
Query: 114 TIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF-DFQKLHKIAIG 172
IG H NLV+L GFC R L+YEYV NG+L++++ G+ + + + KI +G
Sbjct: 866 AIGHVRHKNLVRLLGFCIEGTHRLLIYEYVNNGNLEQWLHGAMRQYGYLTWDARIKILLG 1045
Query: 173 TAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGT 232
TAK +AYLHE + +++H DIK N+L+D KI+DFGLAKL + + T GT
Sbjct: 1046TAKALAYLHEAIEPKVVHRDIKSSNILIDDDFNAKISDFGLAKLLGAGKS-HITTRVMGT 1222
Query: 233 RGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNE 292
GY APE + K DVYSFG+LL E + R D + S ++ W +M N
Sbjct: 1223FGYVAPEYANSGLLNEKSDVYSFGVLLLEAITGRDPVDYNRSAAEVNLVDWL-KMMVGNR 1399
Query: 293 LVVMLALCEIEEKDSEIA-ERMLKVALWCVQYSPNDRPLMSTVVKMLEGE 341
+ IE + S A +R+L AL CV RP MS VV+MLE E
Sbjct: 1400HAEEVVDPNIETRPSTSALKRVLLTALRCVDPDSEKRPKMSQVVRMLESE 1549
>CA916960 weakly similar to GP|14090203|db putative receptor kinase {Oryza
sativa (japonica cultivar-group)}, partial (27%)
Length = 651
Score = 184 bits (467), Expect = 5e-47
Identities = 95/200 (47%), Positives = 133/200 (66%), Gaps = 1/200 (0%)
Frame = +2
Query: 36 KHEFATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVK 95
K + A +E+ + KP R++ ++ IT + +LG GA+G V+KG +S +VAVK
Sbjct: 53 KEKQAIIEKFLEDYRALKPTRYSYVEIKRITNNFGDMLGQGAYGTVYKGSISKEFSVAVK 232
Query: 96 VLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGS 155
+LN + G + F EV T+GR +H+N+V+L GFC KRAL+YE++ NGSL K+I
Sbjct: 233 ILN-VSQGNGQDFLNEVGTMGRIHHVNIVRLIGFCADGFKRALIYEFLPNGSLQKFINSP 409
Query: 156 KNRNDF-DFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLA 214
N+ +F ++KLH+IA+G AKGI YLH+ C RI+H+DIKP+NVLLD PKI DFGLA
Sbjct: 410 DNKKNFLGWKKLHEIALGIAKGIEYLHQGCDQRILHFDIKPQNVLLDHNFIPKICDFGLA 589
Query: 215 KLRSRESNIELNTHFRGTRG 234
KL SR+ +I T RGT G
Sbjct: 590 KLCSRDQSIVSMTAARGTLG 649
>TC80518 weakly similar to PIR|T49986|T49986 lectin-like protein kinase-like
- Arabidopsis thaliana, partial (34%)
Length = 1260
Score = 179 bits (455), Expect = 1e-45
Identities = 116/355 (32%), Positives = 185/355 (51%), Gaps = 12/355 (3%)
Frame = +1
Query: 54 PVRFTPEKLDEITEKYSTI--LGSGAFGVVFKGELSN-GENVAVKVLNCLDMGMEEQFKA 110
P +F E+L T ++ +G+G FG V+KG + + +VA+K ++ +++ +
Sbjct: 10 PKKFIYEELARSTNNFANEHKIGAGGFGAVYKGFIRDLKHHVAIKKVSKESNQGVKEYAS 189
Query: 111 EVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIA 170
EV I + H NLV+LYG+C ++ LVYE+VENGSLD YIF K + + + IA
Sbjct: 190 EVKVISQLRHKNLVQLYGWCHKQNDLLLVYEFVENGSLDSYIF--KGKGLLIWTVRYNIA 363
Query: 171 IGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFR 230
G A + YLHEEC+H ++H DIK NV+LD K+ DFGLA+L + E+ + T
Sbjct: 364 RGLASALLYLHEECEHCVLHRDIKSSNVMLDSNFNTKLGDFGLARLMNHETESK-TTVLA 540
Query: 231 GTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFEN 290
GT GY +PE + + DVYSFG++ EI R+ + S SE + W E++ N
Sbjct: 541 GTYGYLSPEAATRGKASRESDVYSFGVVALEIACGRKAIEPSLSEEHIYLVDWVCELYGN 720
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPF- 349
+L+ E + + ER++ V LWC RP++ VV++L + + + P
Sbjct: 721 GDLLKAADSRLYGEFNEKEVERLMIVGLWCTHTDHLQRPMIRQVVQVLNFDAPLPNLPLQ 900
Query: 350 --------PFHNLVPAKENSTQEGSTADSDTTTSSWRTESSRESGFKTKHNAFEI 396
F+++ + S E + + T++ S T SS+ S AFE+
Sbjct: 901 MNASTYNTSFNSVYSKSKISGFENNQTGTSTSSDSTITGSSQSS------TAFEV 1047
>TC79762 similar to PIR|H96731|H96731 hypothetical protein F5A18.8
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1564
Score = 179 bits (455), Expect = 1e-45
Identities = 111/322 (34%), Positives = 177/322 (54%), Gaps = 4/322 (1%)
Frame = +3
Query: 21 FSST-NNIEVVVQTDSKHEFATMERIFSNINREKPVRFTPEKLDEITEKYSTI--LGSGA 77
FS+T N + V+ D++ E M +RE+ + F+ E L T+ ++ LG G
Sbjct: 258 FSNTLNAVTTQVEGDNEAELQKMA------SREQKI-FSYETLLSATKNFNATHKLGEGG 416
Query: 78 FGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRA 137
FG V+KG+LS+G VAVK L+ +++F E + R H N+V L G+C H ++
Sbjct: 417 FGPVYKGKLSDGREVAVKKLSQTSNQGKKEFMNEAKLLARVQHKNVVNLLGYCVHGTEKI 596
Query: 138 LVYEYVENGSLDKYIF-GSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPE 196
LVYEYV + SLDK++F ++ R D+++ I G AKG+ YLHE+ + IIH DIK
Sbjct: 597 LVYEYVPHESLDKFLFKEAEKREQLDWKRRFGIITGVAKGLLYLHEDSHNCIIHRDIKAS 776
Query: 197 NVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFG 256
N+LLD K KIADFG+A+L E ++ T GT GY APE ++ K DV+S+G
Sbjct: 777 NILLDDKWTAKIADFGMARL-FPEDQSQVKTRVAGTNGYMAPEYMMHGRLSVKADVFSYG 953
Query: 257 ILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKV 316
+L+ E++ +R+ + + W ++M++ + ++ +E + +++
Sbjct: 954 VLVLELITGQRNSSFNLXVEEHNLLDWAYKMYKKGRSLEIVDSALASTVLTEQVDMCIQL 1133
Query: 317 ALWCVQYSPNDRPLMSTVVKML 338
AL C+Q P RP M +V L
Sbjct: 1134ALLCIQGDPQLRPTMRRIVVKL 1199
>TC87048 similar to GP|8778594|gb|AAF79602.1| F5M15.3 {Arabidopsis
thaliana}, partial (77%)
Length = 1571
Score = 179 bits (454), Expect = 2e-45
Identities = 101/274 (36%), Positives = 151/274 (54%), Gaps = 2/274 (0%)
Frame = +1
Query: 68 KYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLY 127
K + ++G G FG VFKG LS GE VAVK L+ ++F EV+ + +H NLVKL
Sbjct: 469 KEANLIGEGGFGKVFKGRLSTGELVAVKQLSHDGRQGFQEFVTEVLMLSLLHHSNLVKLI 648
Query: 128 GFCFHRDKRALVYEYVENGSLDKYIFG-SKNRNDFDFQKLHKIAIGTAKGIAYLHEECKH 186
G+C D+R LVYEY+ GSL+ ++F +++ + KIA+G A+G+ YLH +
Sbjct: 649 GYCTDGDQRLLVYEYMPMGSLEDHLFDLPQDKEPLSWSSRMKIAVGAARGLEYLHCKADP 828
Query: 187 RIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPV 246
+I+ D+K N+LLD PK++DFGLAKL N ++T GT GY APE +
Sbjct: 829 PVIYRDLKSANILLDSDFSPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKL 1008
Query: 247 TYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIE-EK 305
T K D+YSFG++L E++ RR D+S +Q W+ F + V +A ++
Sbjct: 1009TLKSDIYSFGVVLLELITGRRAIDASKKPGEQNLVSWSRPYFSDRRKFVHMADPLLQGHF 1188
Query: 306 DSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLE 339
+ + + C+Q P RPL+ +V LE
Sbjct: 1189PVRCLHQAIAITAMCLQEQPKFRPLIGDIVVALE 1290
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 179 bits (453), Expect = 2e-45
Identities = 110/301 (36%), Positives = 170/301 (55%), Gaps = 16/301 (5%)
Frame = +2
Query: 55 VRFTPEKLDEITEKYST--ILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEV 112
+ F+ +++ EIT +S+ ++G G FG V+K + +G A+K+L E +F+AEV
Sbjct: 152 ILFSYDQILEITNGFSSENVIGEGGFGRVYKALMPDGRVGALKLLKAGSGQGEREFRAEV 331
Query: 113 ITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIG 172
TI R +H +LV L G+C +R L+YE+V NG+LD+++ S+ N D+ K KIAIG
Sbjct: 332 DTISRVHHRHLVSLIGYCIAEQQRVLIYEFVPNGNLDQHLHESQ-WNVLDWPKRMKIAIG 508
Query: 173 TAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGT 232
A+G+AYLHE C +IIH DIK N+LLD E ++ADFGLA+L + ++N ++T GT
Sbjct: 509 AARGLAYLHEGCNPKIIHRDIKSSNILLDDSYEAQVADFGLARL-TDDTNTHVSTRVMGT 685
Query: 233 RGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNE 292
GY APE +T + DV+SFG++L E+V R+ D + + W
Sbjct: 686 FGYMAPEYATSGKLTDRSDVFSFGVVLLELVTGRKPVDPTQPVGDESLVEWARP------ 847
Query: 293 LVVMLALCEIEEKD-SEIAE-------------RMLKVALWCVQYSPNDRPLMSTVVKML 338
+ L IE D SE+A+ RM++ A C+++S RP M + + L
Sbjct: 848 ----ILLRAIETGDFSELADPRLHRQYIDSEMFRMIEAAAACIRHSAPKRPRMVQIARAL 1015
Query: 339 E 339
+
Sbjct: 1016D 1018
>TC83862 similar to PIR|T05270|T05270 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T4L20.80 - Arabidopsis
thaliana, partial (60%)
Length = 948
Score = 176 bits (447), Expect = 1e-44
Identities = 114/309 (36%), Positives = 169/309 (53%), Gaps = 6/309 (1%)
Frame = +1
Query: 57 FTPEKLDEITEKYS--TILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVIT 114
++ ++L+ T+ ++ +++G +G+V++G L +G VAVK L E++FK EV
Sbjct: 13 YSLKELENATDGFAEGSVIGERGYGIVYRGILQDGSIVAVKNLLNNKGQAEKEFKVEVEA 192
Query: 115 IGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNR-NDFDFQKLHKIAIGT 173
IG+ H NLV L G+C KR LVYEYV+NG+L++++ G + + KIA+GT
Sbjct: 193 IGKVRHKNLVGLVGYCAEGAKRMLVYEYVDNGNLEQWLHGDVGPVSPLTWDIRMKIAVGT 372
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
AKG+AYLHE + +++H D+K N+LLD K K++DFGLAKL + + T GT
Sbjct: 373 AKGLAYLHEGLEPKVVHRDVKSSNILLDKKWHAKVSDFGLAKLLGSGKSY-VTTRVMGTF 549
Query: 234 GYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMF---EN 290
GY +PE + DVYSFGILL E+V R D S + ++ W M
Sbjct: 550 GYVSPEYASTGMLNEGSDVYSFGILLMELVTGRSPIDYSRAPAEMNLVDWFKGMVASRRG 729
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP 350
ELV L EI+ + +R L V L C+ N RP M +V MLE + FP
Sbjct: 730 EELVD--PLIEIQPSPRSL-KRALLVCLRCIDLDANKRPKMGQIVHMLEAD------DFP 882
Query: 351 FHNLVPAKE 359
F + + +E
Sbjct: 883 FRSELRTRE 909
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,205,492
Number of Sequences: 36976
Number of extensions: 168660
Number of successful extensions: 2204
Number of sequences better than 10.0: 679
Number of HSP's better than 10.0 without gapping: 1663
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1699
length of query: 402
length of database: 9,014,727
effective HSP length: 98
effective length of query: 304
effective length of database: 5,391,079
effective search space: 1638888016
effective search space used: 1638888016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148970.5


