

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.11 + phase: 0
(382 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
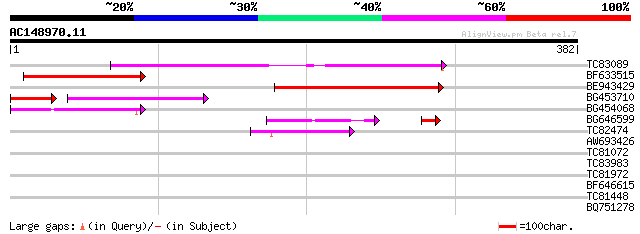
Score E
Sequences producing significant alignments: (bits) Value
TC83089 181 4e-46
BF633515 154 7e-38
BE943429 134 8e-32
BG453710 weakly similar to GP|21537057|gb| unknown {Arabidopsis ... 71 1e-20
BG454068 weakly similar to GP|22202804|dbj hypothetical protein~... 67 9e-12
BG646599 54 4e-09
TC82474 50 2e-06
AW693426 homologue to GP|23307580|dbj similar to serine/threonin... 40 0.001
TC81072 similar to PIR|T05401|T05401 hypothetical protein F10M6.... 31 0.70
TC83983 similar to GP|19386796|dbj|BAB86175. putative alpha-gluc... 30 1.2
TC81972 similar to GP|15290113|dbj|BAB63805. contains ESTs AU031... 29 3.5
BF646615 homologue to PIR|T48395|T483 GT2-like protein - Arabido... 28 4.5
TC81448 weakly similar to OMNI|NT01MC3624 toxin secretion ATP-bi... 28 5.9
BQ751278 similar to PIR|T37665|T37 probable t-complex protein 1 ... 28 7.7
>TC83089
Length = 887
Score = 181 bits (459), Expect = 4e-46
Identities = 103/229 (44%), Positives = 133/229 (57%), Gaps = 3/229 (1%)
Frame = +3
Query: 69 LRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRSNKRIELIY 128
LR++Y YL A L+ ++ + +ELE++RT CV+ YLLYLV CLLFGD+SNKRIEL+Y
Sbjct: 264 LREYYEGYLDSATRLSDPQNLGDPQELEKIRTTCVKWYLLYLVGCLLFGDKSNKRIELVY 443
Query: 129 LTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLDDVCWRPYEEHR 188
LTTM DGYAGMRN+SWG MTLAYLY L++ PG L SVTL+T
Sbjct: 444 LTTMEDGYAGMRNHSWGGMTLAYLYHCLSEVSLPGGGALERSVTLVT------------- 584
Query: 189 EIQDFEEVFWYSGWIMCGVRRVYRHLSERVLRQYGYVQTIPRHPTDVRDLPPPSIVQMFV 248
+GW +C HL ER LR YGYVQT+P PT + L P +V F+
Sbjct: 585 -----------AGWFLC-------HLPERGLR*YGYVQTVPIPPTTIEPLEPAEVVTAFL 710
Query: 249 DFRTHTLKADARGEQAGEDT-WRVADGYVLWYTRVSHPQILPP--IPGD 294
+F H RGE ED + + GY+ + RVSH ++ P +P D
Sbjct: 711 EFALHVQSQQERGESVPEDEGHKHSKGYMK*FYRVSHLLMIAPAAVPED 857
>BF633515
Length = 249
Score = 154 bits (388), Expect = 7e-38
Identities = 72/82 (87%), Positives = 76/82 (91%)
Frame = +2
Query: 10 LDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGYISYPRL 69
LDDVACLTHLPIEGRML HGKKMPKHEGAALLM +LGV+Q EAEKIC QEYGGYISYPRL
Sbjct: 2 LDDVACLTHLPIEGRMLSHGKKMPKHEGAALLMRHLGVSQQEAEKICGQEYGGYISYPRL 181
Query: 70 RDFYTSYLGRANVLAGTEDPEE 91
RDFYT+YLGRAN+LA TEDP E
Sbjct: 182RDFYTTYLGRANLLASTEDPVE 247
>BE943429
Length = 384
Score = 134 bits (336), Expect = 8e-32
Identities = 59/115 (51%), Positives = 76/115 (65%), Gaps = 1/115 (0%)
Frame = +2
Query: 179 VCWRPYEEHREIQDFEEVFWYSGWIMCGVRRVYRHLSERVLRQYGYVQTIPRHPTDVRDL 238
V WRPYE R++ F++V WYSGWIM G +++ RHL ERVLRQYGYVQT+PR PT + L
Sbjct: 23 VIWRPYEHRRDVTPFQDVCWYSGWIMAGKQKMVRHLPERVLRQYGYVQTVPRPPTTIVPL 202
Query: 239 PPPSIVQMFVDFRTHTLKADARGEQAGEDT-WRVADGYVLWYTRVSHPQILPPIP 292
P + F +F H L RG+ ED W+ +DGY+ W+ RVSHP I+ P P
Sbjct: 203 APAEVATAFFEFVVHVLSQQDRGDPVPEDEWWKHSDGYIKWFYRVSHPLIVNPAP 367
>BG453710 weakly similar to GP|21537057|gb| unknown {Arabidopsis thaliana},
partial (6%)
Length = 651
Score = 70.9 bits (172), Expect(2) = 1e-20
Identities = 40/95 (42%), Positives = 54/95 (56%)
Frame = -3
Query: 40 LLMTYLGVAQHEAEKICNQEYGGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVR 99
L+ LG++ A E G+ISYP L+ Y +L A L EE++E R R
Sbjct: 298 LMTRLLGMSDANAMAEIRTESAGHISYPTLKRVYEDHLIEARRLEDPLTREELQERARRR 119
Query: 100 TYCVRCYLLYLVRCLLFGDRSNKRIELIYLTTMAD 134
+CVR LLYLV C+LF ++N+ I+LIYL MAD
Sbjct: 118 QWCVRSLLLYLVDCVLFTYKTNRHIDLIYLDCMAD 14
Score = 46.6 bits (109), Expect(2) = 1e-20
Identities = 20/31 (64%), Positives = 25/31 (80%)
Frame = -1
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKK 31
+PMGEMT+TLDDV L H+PI G MLDH ++
Sbjct: 414 LPMGEMTITLDDVYNLLHIPIHGCMLDHDEQ 322
>BG454068 weakly similar to GP|22202804|dbj hypothetical protein~predicted by
GeneMark.hmm etc. {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 673
Score = 67.4 bits (163), Expect = 9e-12
Identities = 36/94 (38%), Positives = 52/94 (55%), Gaps = 3/94 (3%)
Frame = +3
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P+GEMT+ LDD++CL H+P+ G +L H + + H+G L+ YLG+ E +
Sbjct: 387 LPVGEMTIALDDISCLLHIPVGGNLLFH-ESLSIHQGTEYLVNYLGLEFEEIAAETKRLK 563
Query: 61 GGYISYPRLRDFYTSYLGRANVLA---GTEDPEE 91
+I+Y L YTSYL A A G ED E
Sbjct: 564 SAHITYDTLLSIYTSYLTEAKSYANQPGEEDSME 665
>BG646599
Length = 711
Score = 54.3 bits (129), Expect(2) = 4e-09
Identities = 30/76 (39%), Positives = 43/76 (56%)
Frame = +3
Query: 174 LTLDDVCWRPYEEHREIQDFEEVFWYSGWIMCGVRRVYRHLSERVLRQYGYVQTIPRHPT 233
LT +DV W P++ HR++ F+++ SG+I V +L ER LRQ+GY+Q IPR
Sbjct: 219 LTPNDVIWCPFDSHRQVIPFDDICLSSGYIR-WCSNVVPYLPERCLRQFGYIQYIPR--- 386
Query: 234 DVRDLPPPSIVQMFVD 249
PPP+ VD
Sbjct: 387 -----PPPNFNTFNVD 419
Score = 23.9 bits (50), Expect(2) = 4e-09
Identities = 7/13 (53%), Positives = 10/13 (76%)
Frame = +1
Query: 278 WYTRVSHPQILPP 290
WY +VSHP ++ P
Sbjct: 517 WYYKVSHPHLIRP 555
>TC82474
Length = 992
Score = 49.7 bits (117), Expect = 2e-06
Identities = 24/73 (32%), Positives = 40/73 (53%), Gaps = 3/73 (4%)
Frame = -2
Query: 163 GHRVLGGSVTLLT---LDDVCWRPYEEHREIQDFEEVFWYSGWIMCGVRRVYRHLSERVL 219
GH+ +T L +DDV + Y+ H + F+++ + WI C + Y HL E V+
Sbjct: 838 GHKSPNNYMTFLDRVEVDDVSFNQYDFHHQTCPFDDIS*FFRWIECDRKMNYHHLLELVM 659
Query: 220 RQYGYVQTIPRHP 232
+ YG++Q+ RHP
Sbjct: 658 K*YGHIQSTLRHP 620
>AW693426 homologue to GP|23307580|dbj similar to serine/threonine
phosphatase PP7 {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 209
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/69 (34%), Positives = 34/69 (48%)
Frame = +1
Query: 106 YLLYLVRCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHR 165
+LL LV C LF D+S + + YL D Y+WG LA+LY L A +
Sbjct: 1 FLLCLVGCTLFSDKSTFVVNVAYLEFFRD-LDSCGGYAWGVAALAHLYDNLRYASFHHMK 177
Query: 166 VLGGSVTLL 174
+ G +TL+
Sbjct: 178 SISGYLTLI 204
>TC81072 similar to PIR|T05401|T05401 hypothetical protein F10M6.90 -
Arabidopsis thaliana, partial (13%)
Length = 745
Score = 31.2 bits (69), Expect = 0.70
Identities = 11/39 (28%), Positives = 26/39 (66%), Gaps = 3/39 (7%)
Frame = +2
Query: 80 ANVLAGTEDPEEVEELERV---RTYCVRCYLLYLVRCLL 115
A+++ ++ P + +R+ +T+C+RC L+Y++RC +
Sbjct: 302 ASIIGSSQKPNR*QTFQRLCYDQTWCLRCNLVYVLRCFV 418
>TC83983 similar to GP|19386796|dbj|BAB86175. putative alpha-glucosidase 1
{Oryza sativa (japonica cultivar-group)}, partial (11%)
Length = 656
Score = 30.4 bits (67), Expect = 1.2
Identities = 13/60 (21%), Positives = 27/60 (44%), Gaps = 5/60 (8%)
Frame = -2
Query: 282 VSHPQILPPIPGDLLRPANEEQI-----IAEQWQRYEARNSPDTYDMVSGAVAYADAQLG 336
+SHP++ D+ RP+++ + I E W + R+ D + + A+ + G
Sbjct: 562 LSHPRVYKHASSDIYRPSHQSTLIQPSKIVESWHEFRDRDKDDMFPNTNSLCAHLETDRG 383
>TC81972 similar to GP|15290113|dbj|BAB63805. contains ESTs AU031749(R1464)
D24170(R1464)~unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (10%)
Length = 684
Score = 28.9 bits (63), Expect = 3.5
Identities = 27/120 (22%), Positives = 49/120 (40%)
Frame = +2
Query: 208 RRVYRHLSERVLRQYGYVQTIPRHPTDVRDLPPPSIVQMFVDFRTHTLKADARGEQAGED 267
RR+ + S+R+ G++QT+P PTD ++ P S + THT + ++
Sbjct: 83 RRILQQGSDRLAFIKGHIQTLP--PTDFQN-PDASETSI-----THTPTFPEQQHTQTDE 238
Query: 268 TWRVADGYVLWYTRVSHPQILPPIPGDLLRPANEEQIIAEQWQRYEARNSPDTYDMVSGA 327
D + H Q+LP D +Q AE+ + + N D + + +
Sbjct: 239 ILSFPDSEIQPEPEPEHVQLLPQTLPDSFNEIPRQQTRAEEPRSFNFINPSDVSNAIDAS 418
>BF646615 homologue to PIR|T48395|T483 GT2-like protein - Arabidopsis
thaliana, partial (16%)
Length = 654
Score = 28.5 bits (62), Expect = 4.5
Identities = 16/41 (39%), Positives = 25/41 (60%), Gaps = 5/41 (12%)
Frame = -2
Query: 13 VACLTHLPIEG----RMLDHGKKMPKHEGA-ALLMTYLGVA 48
V+CL HLP+E + + K+P +EG AL++ +GVA
Sbjct: 365 VSCLGHLPVEASTAMKAVVSASKLPLNEGVEALVVVLVGVA 243
>TC81448 weakly similar to OMNI|NT01MC3624 toxin secretion ATP-binding
protein putative {Magnetococcus sp. MC-1}, partial (4%)
Length = 655
Score = 28.1 bits (61), Expect = 5.9
Identities = 20/87 (22%), Positives = 39/87 (43%), Gaps = 3/87 (3%)
Frame = +3
Query: 299 ANEEQIIAEQWQRYEARNSPDTYDMVSGAVAYADAQLGQEEVMSMTPQQSFEAITHMREQ 358
A +E++ + + E + +V + A +L +E + + Q EA M+E+
Sbjct: 129 ATQEKLQHQLQEATEKLQHQEALQLVKHQLQEATEKLQHQEALQLVKHQHQEA---MQEK 299
Query: 359 IAPILTRRRAQRP---RRRHHHQDQDQ 382
+ L ++ + + RHHHQ Q Q
Sbjct: 300 LPQALHQQEVHQE*HHQVRHHHQQQHQ 380
>BQ751278 similar to PIR|T37665|T37 probable t-complex protein 1 epsilon
subunit - fission yeast (Schizosaccharomyces pombe),
partial (26%)
Length = 596
Score = 27.7 bits (60), Expect = 7.7
Identities = 21/71 (29%), Positives = 31/71 (43%), Gaps = 1/71 (1%)
Frame = +1
Query: 311 RYEARNSPDTYDMV-SGAVAYADAQLGQEEVMSMTPQQSFEAITHMREQIAPILTRRRAQ 369
R E R P Y SG A Q ++E S P + H ++ + P +R+
Sbjct: 115 RDEGRAGPSLYRRQRSGKEEEAIWQRSRQEPHSRRP----DCREHCQDLLGPPRSRQDPH 282
Query: 370 RPRRRHHHQDQ 380
PRRRHH ++
Sbjct: 283 FPRRRHHSDER 315
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,689,395
Number of Sequences: 36976
Number of extensions: 180881
Number of successful extensions: 900
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 886
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 895
length of query: 382
length of database: 9,014,727
effective HSP length: 98
effective length of query: 284
effective length of database: 5,391,079
effective search space: 1531066436
effective search space used: 1531066436
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148970.11


