

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.6 + phase: 0
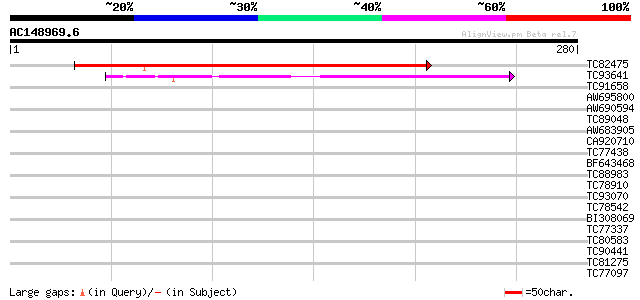
(280 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82475 similar to GP|20146289|dbj|BAB89071. peptide deformylase... 176 7e-45
TC93641 similar to SP|Q9FV54|DEFC_LYCES Peptide deformylase chl... 87 5e-18
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 36 0.019
AW695800 similar to GP|13357246|gb| unknown protein {Oryza sativ... 32 0.27
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 32 0.27
TC89048 weakly similar to PIR|T46215|T46215 hypothetical protein... 31 0.46
AW683905 weakly similar to GP|9757854|dbj| contains similarity t... 30 0.79
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 30 0.79
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 30 1.0
BF643468 weakly similar to PIR|F96682|F96 hypothetical protein F... 30 1.0
TC88983 similar to SP|P93231|VP41_LYCES Vacuolar assembly protei... 30 1.3
TC78910 similar to GP|20260424|gb|AAM13110.1 putative protein {A... 29 1.8
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 29 1.8
TC78542 weakly similar to PIR|T05059|T05059 hypothetical protein... 29 2.3
BI308069 similar to GP|15451228|gb photoreceptor-interacting pro... 29 2.3
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 29 2.3
TC80583 squalene monooxygenase 1 [Medicago truncatula] 29 2.3
TC90441 similar to PIR|T00872|T00872 probable protein kinase At2... 28 3.0
TC81275 similar to PIR|E84833|E84833 hypothetical protein At2g40... 28 3.0
TC77097 similar to PIR|T05572|T05572 glucose-6-phosphate isomera... 28 3.0
>TC82475 similar to GP|20146289|dbj|BAB89071. peptide deformylase-like
protein {Oryza sativa (japonica cultivar-group)},
partial (52%)
Length = 654
Score = 176 bits (447), Expect = 7e-45
Identities = 103/187 (55%), Positives = 124/187 (66%), Gaps = 11/187 (5%)
Frame = +3
Query: 33 VFSNATVSSSSSSCNTETPPSNTKLSSFSSASS-----------ETAFLSKTLKSELPHI 81
+F+ T + S S ++ PP LSS SS ++ T K K +LP
Sbjct: 93 LFTLQTTTPFSLSPFSKPPPLTLTLSSSSSQNATIRTRAGFFFGRTKDDKKKKKMDLPDT 272
Query: 82 VQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLEE 141
V+AGDPVLHEPA+EVD SEI SDK+QKIID MI VMR APG+ L+A +IG+ RIIVLE+
Sbjct: 273 VKAGDPVLHEPAQEVDPSEIMSDKVQKIIDDMIRVMRKAPGVGLAAPQIGVSSRIIVLED 452
Query: 142 PKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYLD 201
+E + +EV K DR PFDLLVILNPKLK S +T LFFEGCLSV GF AVVER+LD
Sbjct: 453 TEEFISYAPKEVLKAQDRHPFDLLVILNPKLKSTSKRTALFFEGCLSVDGFXAVVERHLD 632
Query: 202 VEVEGFD 208
VEV G D
Sbjct: 633 VEVTGLD 653
>TC93641 similar to SP|Q9FV54|DEFC_LYCES Peptide deformylase chloroplast
precursor (EC 3.5.1.88) (PDF) (Polypeptide deformylase).
[Tomato], partial (59%)
Length = 644
Score = 87.4 bits (215), Expect = 5e-18
Identities = 63/203 (31%), Positives = 106/203 (52%), Gaps = 1/203 (0%)
Frame = +1
Query: 48 TETPPSNTKLSSFSSASSETAFLSKTLKSELP-HIVQAGDPVLHEPAREVDHSEINSDKI 106
T + S TK S+FS++ +E A L L+ E P I + DP L + + + + D +
Sbjct: 49 TLSSSSRTK-SAFSASQNEFASLGD-LEFEAPLKIAKYPDPKLRKKNKRIGTFD---DNL 213
Query: 107 QKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLEEPKENLYNYTEEVNKIIDRRPFDLLV 166
+K++D M VM GI LSA ++GI ++++V N + +R + +V
Sbjct: 214 KKLVDEMFDVMYETDGIGLSAPQVGINVQLMVF--------------NPVGERGEGEEIV 351
Query: 167 ILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYLDVEVEGFDRYGEPIKINASGWHARIL 226
++NP++ +S ++ EGCLS + V+R V+++ D G+ ++ SG ARI
Sbjct: 352 LVNPRVGKRSLNRTIYNEGCLSFPRIRGDVKRPEYVKIDALDVKGKRFSVSLSGLPARIF 531
Query: 227 QHECDHLDGTLYVDKMVPRTFRS 249
QHE DHL G L+ ++M F S
Sbjct: 532 QHEFDHLQGXLFFERMADEVFDS 600
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 35.8 bits (81), Expect = 0.019
Identities = 22/59 (37%), Positives = 38/59 (64%), Gaps = 4/59 (6%)
Frame = -2
Query: 25 LRVVPMMLVF----SNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELP 79
L ++P++L+ S+++ SSSSSS ++ + S++ SS SS+SS T+ SK+L P
Sbjct: 222 LALLPLLLLLFPFPSSSSSSSSSSSSSSSSSSSSSSSSSPSSSSSSTSISSKSLSPASP 46
>AW695800 similar to GP|13357246|gb| unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (22%)
Length = 430
Score = 32.0 bits (71), Expect = 0.27
Identities = 20/68 (29%), Positives = 33/68 (48%)
Frame = +1
Query: 26 RVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPHIVQAG 85
R+ P +FS+ ++SSS SSC+T SN KL+ +S ++ ++ +
Sbjct: 91 RMPPSCSMFSHVSMSSSPSSCSTFNSKSNNKLNWTPIRASSEGLPNELVEDSKFVPINVE 270
Query: 86 DPVLHEPA 93
DP PA
Sbjct: 271 DPRYGPPA 294
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 32.0 bits (71), Expect = 0.27
Identities = 17/45 (37%), Positives = 29/45 (63%)
Frame = -3
Query: 35 SNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELP 79
S+++ SSSSSSC++ + S++ SS S +SS ++ S + S P
Sbjct: 205 SSSSSSSSSSSCSSSSSSSSSSSSSSSCSSSSSSSCSSSSSSTSP 71
Score = 31.2 bits (69), Expect = 0.46
Identities = 16/41 (39%), Positives = 27/41 (65%)
Frame = -3
Query: 35 SNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLK 75
S+++ SSSSSSC++ + S + SS +S SS ++ S T +
Sbjct: 157 SSSSSSSSSSSCSSSSSSSCSSSSSSTSPSSSSSLCSSTFR 35
Score = 28.9 bits (63), Expect = 2.3
Identities = 17/44 (38%), Positives = 27/44 (60%)
Frame = -3
Query: 35 SNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSEL 78
S+++ SSSSSS ++ + S+ SS SS SS ++ S + S L
Sbjct: 184 SSSSCSSSSSSSSSSSSSSSCSSSSSSSCSSSSSSTSPSSSSSL 53
Score = 28.1 bits (61), Expect = 3.9
Identities = 16/47 (34%), Positives = 30/47 (63%)
Frame = -3
Query: 30 MMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKS 76
++L S+++ SSSSSS ++ + S++ SS S+SS ++ S + S
Sbjct: 265 LILSISSSSPSSSSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSS 125
Score = 27.7 bits (60), Expect = 5.1
Identities = 16/46 (34%), Positives = 30/46 (64%)
Frame = -3
Query: 28 VPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKT 73
V ++ + S++ SSSSSS ++ + S++ SS SS+SS ++ S +
Sbjct: 268 VLILSISSSSPSSSSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSS 131
Score = 26.9 bits (58), Expect = 8.7
Identities = 16/42 (38%), Positives = 27/42 (64%)
Frame = -3
Query: 35 SNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKS 76
S+++ SSSSSS ++ + S + SS SS+SS ++ S + S
Sbjct: 229 SSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSSCSSSSSS 104
Score = 26.9 bits (58), Expect = 8.7
Identities = 16/48 (33%), Positives = 30/48 (62%)
Frame = -3
Query: 26 RVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKT 73
RV+ + + S+ + SSSSSS ++ + S++ S SS+SS ++ S +
Sbjct: 271 RVLILSISSSSPSSSSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSS 128
>TC89048 weakly similar to PIR|T46215|T46215 hypothetical protein T8P19.220
- Arabidopsis thaliana, partial (35%)
Length = 1329
Score = 31.2 bits (69), Expect = 0.46
Identities = 18/43 (41%), Positives = 26/43 (59%)
Frame = -1
Query: 34 FSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKS 76
FS+ SSSSSS ++ +PPS++ S SS S + F S + S
Sbjct: 537 FSSPASSSSSSSASSSSPPSSSSSLSESSCPSISFFSSDSATS 409
>AW683905 weakly similar to GP|9757854|dbj| contains similarity to apoptosis
antagonizing transcription factor~gene_id:MFB13.10,
partial (12%)
Length = 492
Score = 30.4 bits (67), Expect = 0.79
Identities = 17/41 (41%), Positives = 28/41 (67%), Gaps = 3/41 (7%)
Frame = -3
Query: 34 FSNATVSSSS---SSCNTETPPSNTKLSSFSSASSETAFLS 71
FS+ ++SSS SSC + + PS++ SSFSS++S ++ S
Sbjct: 292 FSSCSISSSLHPFSSCPSSSSPSSSPSSSFSSSNSLSSLTS 170
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 30.4 bits (67), Expect = 0.79
Identities = 15/45 (33%), Positives = 29/45 (64%)
Frame = +1
Query: 30 MMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTL 74
++++ N +SSSSS ++ + ++ SSFSS+SS +A S ++
Sbjct: 253 LIIIKKNPPATSSSSSSSSSSSSFSSSSSSFSSSSSSSASASASI 387
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 - garden
pea, partial (93%)
Length = 1607
Score = 30.0 bits (66), Expect = 1.0
Identities = 19/51 (37%), Positives = 31/51 (60%)
Frame = -3
Query: 30 MMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPH 80
++LV S+++ SSSSSS ++ + S+ SS SS+SS + S S + H
Sbjct: 1143 LVLVSSSSSSSSSSSSSSSSSSSSSASSSSSSSSSSRSPNSSP*AASPVNH 991
Score = 28.1 bits (61), Expect = 3.9
Identities = 23/67 (34%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Frame = -3
Query: 28 VPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPHIVQ-AGD 86
+P+ L V SSSS ++ + S++ SS SSASS + S + S P+ A
Sbjct: 1173 IPLFLADDFFLVLVSSSSSSSSSSSSSSSSSSSSSASSSS---SSSSSSRSPNSSP*AAS 1003
Query: 87 PVLHEPA 93
PV H+ A
Sbjct: 1002 PVNHDTA 982
>BF643468 weakly similar to PIR|F96682|F96 hypothetical protein F12P19.2
[imported] - Arabidopsis thaliana, partial (33%)
Length = 655
Score = 30.0 bits (66), Expect = 1.0
Identities = 15/43 (34%), Positives = 28/43 (64%)
Frame = -2
Query: 38 TVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPH 80
T ++S++S TE+PP ++ + SFS S+ T +S + S +P+
Sbjct: 540 TTTASNTS*LTESPPLDSTIHSFSFLSTFTTSVSNRVNS*IPN 412
>TC88983 similar to SP|P93231|VP41_LYCES Vacuolar assembly protein VPS41
homolog. [Tomato] {Lycopersicon esculentum}, partial
(17%)
Length = 668
Score = 29.6 bits (65), Expect = 1.3
Identities = 20/56 (35%), Positives = 31/56 (54%)
Frame = -3
Query: 10 LFETEMEAARRLASRLRVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASS 65
L TE + A +A+ +P +L + + SSSSSS + + S++ SS SS SS
Sbjct: 315 LSATETQDAASVANNEGTLPPIL*YFSLGSSSSSSSSTSSSSSSSSSSSSSSSRSS 148
>TC78910 similar to GP|20260424|gb|AAM13110.1 putative protein {Arabidopsis
thaliana}, partial (40%)
Length = 1216
Score = 29.3 bits (64), Expect = 1.8
Identities = 23/90 (25%), Positives = 38/90 (41%), Gaps = 4/90 (4%)
Frame = +1
Query: 51 PPSNTKLSSFSSASSETAFLSKTLKSELPHIVQAGDPVLHEPAREVDHSEINSDKIQKII 110
PP+ + S S+ S T P L AR VD ++ +Q ++
Sbjct: 766 PPAGSGFISGFSSMSRRYSADSTPNLSFPF---GPSSYLSAQARVVDEYTMSQTILQNML 936
Query: 111 DG----MILVMRNAPGISLSAQKIGIPLRI 136
DG M++V+ A ++ A+ G+P RI
Sbjct: 937 DGGATGMLIVVTGASHVTYGARGTGVPARI 1026
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 29.3 bits (64), Expect = 1.8
Identities = 17/42 (40%), Positives = 26/42 (61%)
Frame = -3
Query: 36 NATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSE 77
+A SSSSSS ++ + S++ SS SS+SS + +S SE
Sbjct: 347 SANPSSSSSSSSSSSSSSSSSSSSSSSSSSSSKSISSISSSE 222
Score = 26.9 bits (58), Expect = 8.7
Identities = 14/33 (42%), Positives = 24/33 (72%)
Frame = -3
Query: 35 SNATVSSSSSSCNTETPPSNTKLSSFSSASSET 67
+N + SSSSSS ++ + S++ SS SS+SS++
Sbjct: 344 ANPSSSSSSSSSSSSSSSSSSSSSSSSSSSSKS 246
>TC78542 weakly similar to PIR|T05059|T05059 hypothetical protein M3E9.120 -
Arabidopsis thaliana, partial (2%)
Length = 2075
Score = 28.9 bits (63), Expect = 2.3
Identities = 36/167 (21%), Positives = 68/167 (40%), Gaps = 22/167 (13%)
Frame = +1
Query: 14 EMEAARRLASRLRVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKT 73
E ++A+ ++ + + L F + +SS SS+ T S++ SS L
Sbjct: 1150 EDDSAKCSSASAGITAVDLGFHKSFLSSHSSADETNQSASSSLEFQISSLKQHVNLLESK 1329
Query: 74 LKS-------------ELPHIVQAGDPVLHEPAREVDHSEINSD-------KIQKIIDGM 113
L EL + +G E A + + E+ + K++ ++ +
Sbjct: 1330 LGELQGVLSSKDSRIVELETALSSGKFPKEESANTLGYKEVEYEIEDLFRQKVEAEVEYL 1509
Query: 114 IL--VMRNAPGISLSAQKIGIPLRIIVLEEPKENLYNYTEEVNKIID 158
+ VM+N K+G L+ +LEE ++ N + +NKIID
Sbjct: 1510 AIAKVMQNL--------KVGADLQFTLLEEQEKLSENQAQVLNKIID 1626
>BI308069 similar to GP|15451228|gb photoreceptor-interacting protein-like
{Arabidopsis thaliana}, partial (19%)
Length = 522
Score = 28.9 bits (63), Expect = 2.3
Identities = 20/66 (30%), Positives = 28/66 (42%)
Frame = +2
Query: 12 ETEMEAARRLASRLRVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLS 71
E M A + LRVV +L F S++SSS T TP + + S + S
Sbjct: 248 EACMHAVQNERLPLRVVVQVLFFEQMRASTTSSSGGTSTPDLPGSIRALHPGGSHGSSRS 427
Query: 72 KTLKSE 77
T +E
Sbjct: 428 TTTNTE 445
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 28.9 bits (63), Expect = 2.3
Identities = 14/33 (42%), Positives = 24/33 (72%)
Frame = -2
Query: 35 SNATVSSSSSSCNTETPPSNTKLSSFSSASSET 67
S++ SSS+ S ++ +PPS++ SS SS+SS +
Sbjct: 724 SSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSS 626
Score = 28.5 bits (62), Expect = 3.0
Identities = 17/58 (29%), Positives = 31/58 (53%)
Frame = -2
Query: 35 SNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPHIVQAGDPVLHEP 92
++++ SSSSSS ++ + PS++ S SS+SS ++ S + P + P P
Sbjct: 748 ASSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSPFSSAP 575
Score = 26.9 bits (58), Expect = 8.7
Identities = 17/45 (37%), Positives = 23/45 (50%)
Frame = -2
Query: 32 LVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKS 76
L F SSSSSS ++ S+T SS SS S ++ S + S
Sbjct: 769 LFFGGCKASSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSS 635
>TC80583 squalene monooxygenase 1 [Medicago truncatula]
Length = 1830
Score = 28.9 bits (63), Expect = 2.3
Identities = 17/65 (26%), Positives = 27/65 (41%)
Frame = -3
Query: 37 ATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPHIVQAGDPVLHEPAREV 96
+T SSS SS N PP + + S S + + T + LP + +P P +
Sbjct: 463 STQSSSPSSINLR*PPGCSNSPTIRSGSFRSLSIISTRRPSLPRV*ARAEPATPAPTMMI 284
Query: 97 DHSEI 101
S +
Sbjct: 283 STSPL 269
>TC90441 similar to PIR|T00872|T00872 probable protein kinase At2g45590
[imported] - Arabidopsis thaliana, partial (27%)
Length = 935
Score = 28.5 bits (62), Expect = 3.0
Identities = 18/67 (26%), Positives = 31/67 (45%)
Frame = -2
Query: 141 EPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYL 200
EPK NYT +++IID + +++ + NK FL + C H +V Y
Sbjct: 415 EPKAEHRNYTHHISRIIDH---------HHRIQEQCNKQFLSYSVCYLFH-HSSVYHHYP 266
Query: 201 DVEVEGF 207
+++ F
Sbjct: 265 NLDYFSF 245
>TC81275 similar to PIR|E84833|E84833 hypothetical protein At2g40760
[imported] - Arabidopsis thaliana, partial (27%)
Length = 883
Score = 28.5 bits (62), Expect = 3.0
Identities = 17/45 (37%), Positives = 29/45 (63%)
Frame = +3
Query: 32 LVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKS 76
L + +SSSSSS ++ TP S T+ +++SS+S +F + +L S
Sbjct: 9 LSLTTRMLSSSSSSSSSTTPSSETR-TAYSSSSPNPSFKNLSLFS 140
>TC77097 similar to PIR|T05572|T05572 glucose-6-phosphate isomerase (EC
5.3.1.9) - Arabidopsis thaliana, partial (40%)
Length = 1017
Score = 28.5 bits (62), Expect = 3.0
Identities = 19/60 (31%), Positives = 30/60 (49%)
Frame = -1
Query: 5 QGSSSLFETEMEAARRLASRLRVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSAS 64
+G L ET A ++ASR+ + ++ A + S S C T P S ++ FSS+S
Sbjct: 252 EGGLILPETISSAKPQIASRVLSICVLRNIFGARLESLSQ*CPTIRPSSGFAIAPFSSSS 73
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,745,694
Number of Sequences: 36976
Number of extensions: 107253
Number of successful extensions: 1214
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 989
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1101
length of query: 280
length of database: 9,014,727
effective HSP length: 95
effective length of query: 185
effective length of database: 5,502,007
effective search space: 1017871295
effective search space used: 1017871295
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148969.6


