

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.3 + phase: 2 /pseudo
(595 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
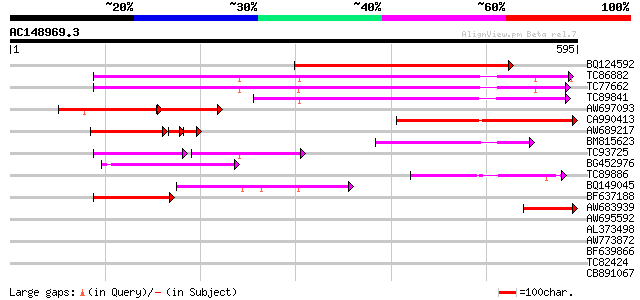
Score E
Sequences producing significant alignments: (bits) Value
BQ124592 similar to GP|20466159|gb putative membrane transporter... 390 e-109
TC86882 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabido... 291 4e-79
TC77662 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabido... 283 1e-76
TC89841 similar to PIR|T02307|T02307 probable membrane transport... 207 1e-53
AW697093 similar to GP|20466159|gb putative membrane transporter... 114 9e-52
CA990413 similar to PIR|T05632|T05 hypothetical protein F20D10.1... 199 2e-51
AW689217 similar to GP|15144504|gb| putative permease {Lycopersi... 100 2e-27
BM815623 similar to GP|14334908|gb putative membrane transporter... 118 6e-27
TC93725 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabido... 69 9e-24
BG452976 weakly similar to GP|15983805|gb At1g49960/F2J10_14 {Ar... 98 8e-21
TC89886 weakly similar to GP|14587294|dbj|BAB61205. putative per... 82 4e-16
BQ149045 similar to PIR|T02307|T02 probable membrane transporter... 79 4e-15
BF637188 similar to GP|14334908|gb putative membrane transporter... 73 3e-13
AW683939 similar to GP|20466159|gb| putative membrane transporte... 65 7e-11
AW695592 weakly similar to PIR|T00984|T009 probable membrane tra... 33 0.40
AL373498 similar to PIR|D84651|D84 hypothetical protein At2g2568... 31 1.5
AW773872 similar to GP|21595706|gb unknown {Arabidopsis thaliana... 30 2.0
BF639866 homologue to GP|3036792|emb| putative protein (fragment... 30 2.6
TC82424 similar to PIR|T51283|T51283 glucan 1 3-beta-glucosidase... 30 3.4
CB891067 similar to PIR|C86334|C863 hypothetical protein AAF7990... 29 5.8
>BQ124592 similar to GP|20466159|gb putative membrane transporter
{Arabidopsis thaliana}, partial (32%)
Length = 690
Score = 390 bits (1003), Expect = e-109
Identities = 192/229 (83%), Positives = 205/229 (88%)
Frame = +2
Query: 300 MKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLL 359
MK+C+V+TS + S PWF+FPYPLQWGTPVF+WKMA+VMCVVSLISSVDSVG+YH SSLL
Sbjct: 2 MKYCRVDTSHAIKSSPWFKFPYPLQWGTPVFHWKMALVMCVVSLISSVDSVGSYHASSLL 181
Query: 360 AASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGAC 419
AS PPTPGVLSRGIGLEG SS+LAGLWGTG GSTTLTENVHTIA TKMGSRR VQLGAC
Sbjct: 182 VASRPPTPGVLSRGIGLEGLSSVLAGLWGTGTGSTTLTENVHTIAVTKMGSRRAVQLGAC 361
Query: 420 LLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSL 479
+LIVLSL GKVGGFIASIP MVAGLLC MWAMLTALGLSNLRY+E GSSRNIII+GLSL
Sbjct: 362 ILIVLSLVGKVGGFIASIPGVMVAGLLCFMWAMLTALGLSNLRYSEAGSSRNIIIIGLSL 541
Query: 480 FFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVL 528
FFSLSIPAYFQQY SP SN SVPSYFQPYIV SHGPF+SKY LNY L
Sbjct: 542 FFSLSIPAYFQQYGISPNSNLSVPSYFQPYIVASHGPFQSKYGGLNYFL 688
>TC86882 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabidopsis
thaliana}, complete
Length = 2140
Score = 291 bits (746), Expect = 4e-79
Identities = 170/528 (32%), Positives = 276/528 (52%), Gaps = 25/528 (4%)
Frame = +2
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
++Y +T P + G QHYL ++G+ +L P + P MG ++E A ++ T+L V+G+
Sbjct: 350 VSYCITSPPPWPEAILLGFQHYLVMLGTTVLIPTSLVPQMGGGNEEKAKVIQTLLFVAGI 529
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNE--NKFKHIMKELQGAIIIGS 206
TL+ T+FGSRLP + G S+ ++ ++II + F + + K K IM+ QGA+I+ S
Sbjct: 530 NTLVQTLFGSRLPAVIGGSYTFVPATISIILAGRFNDEPDPIEKIKKIMRATQGALIVAS 709
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLLFF 260
Q +LG++GL + RF++P+ ++ VG F +A+ + L+F
Sbjct: 710 TLQIVLGFSGLWRNVARFLSPLSAVPLVSLVGFGLYELGFPGVAKCVEIGLPELVLLVFV 889
Query: 261 VLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMT 312
++ + G H+F ++V +A+ W +AF+LT G H C+ ++S +
Sbjct: 890 SQFVPHVLHSGKHVFDRFSVLFTVAIVWLYAFILTVGGAYNHVKRTTQMTCRTDSSGLID 1069
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
+ PW R PYP QWG P F+ A M + S ++ V+S G + A++ P P +LSR
Sbjct: 1070AAPWIRVPYPFQWGAPSFDAGEAFAMMMTSFVALVESSGAFIAVYRFASATPLPPSILSR 1249
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G LL+GL+GTG+GS+ EN +A T++GSRR VQ+ A +I S+ GK G
Sbjct: 1250GIGWQGVGILLSGLFGTGIGSSVSVENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGA 1429
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP +VA L C+ +A + + GLS L++ S R ++G S+F LSIP YF +Y
Sbjct: 1430VFASIPPPIVAALYCLFFAYVGSGGLSFLQFCNLNSFRTKFVLGFSIFLGLSIPQYFNEY 1609
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ + GP + N ++N+ F +A +VA LDNT+
Sbjct: 1610TA----------------INGFGPVHTGARWFNDIVNVPFQSKAFVAGVVAYFLDNTLHK 1741
Query: 551 --PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF-----RWV 591
+++R + W K + D Y LP + + F RW+
Sbjct: 1742KESAIRKDRGKHWWDKYRSFKTDTRSEEFYSLPFNLNKYFPSV*LRWL 1885
>TC77662 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabidopsis
thaliana}, complete
Length = 1953
Score = 283 bits (725), Expect = 1e-76
Identities = 168/520 (32%), Positives = 270/520 (51%), Gaps = 20/520 (3%)
Frame = +2
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
++Y +T P + G QH+L ++G+ +L P + P MG + E A ++ T+L V+G+
Sbjct: 176 ISYCITSPPPWPEAILLGFQHFLVMLGTTVLIPTALVPQMGGGNAEKAKVIETLLFVAGI 355
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNE--NKFKHIMKELQGAIIIGS 206
TL+ T+FGSRLP + G S+ Y+ ++II + F + KFK IM+ +QGA+I+ S
Sbjct: 356 NTLVQTLFGSRLPAVIGGSYTYVPTTISIILAGRFSNEPDPIEKFKKIMRAVQGALIVAS 535
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLLFF 260
Q +LG++GL + RF++P+ ++ VG F +A+ + L+F
Sbjct: 536 TLQIVLGFSGLWRNVARFLSPLSAVPLVSLVGFGLYELGFPGVAKCVEIGLPELILLVFV 715
Query: 261 VLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMK--------HCQVNTSDTMT 312
Y+ + G +IF ++V +A+ W +A LLT G C+ + + +
Sbjct: 716 SQYVPHVLHSGKNIFDRFSVLFTIAIVWIYAVLLTVGGAYNGSPPKTQTSCRTDRAGLID 895
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
+ PW R PYP QWG P F+ A M + S ++ V+S G + A++ P P +LSR
Sbjct: 896 AAPWIRVPYPFQWGAPTFDAGEAFAMMMASFVALVESSGAFIAVYRFASATPLPPSILSR 1075
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G LL+GL+GT GS+ EN +A T++GSRR VQ+ A +I S+ GK G
Sbjct: 1076GIGWQGVGILLSGLFGTISGSSVSVENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGA 1255
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP A++A L C+ +A + A GLS L++ S R I+G S+F LS+P YF +Y
Sbjct: 1256VFASIPPAIIAALYCLFFAYVGAGGLSFLQFCNLNSFRTKFILGFSIFLGLSVPQYFNEY 1435
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ + +GP + N ++N+ F +A +VA LDNT+
Sbjct: 1436TA----------------INGYGPVHTGGRWFNDMVNVPFQSKAFVAGVVAYFLDNTLHK 1567
Query: 551 --PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
+++R + W K + D Y LP + + F
Sbjct: 1568RDSSIRKDRGKHWWDKYKSFKGDTRSEEFYSLPFNLNKYF 1687
>TC89841 similar to PIR|T02307|T02307 probable membrane transporter
At2g34190 [imported] - Arabidopsis thaliana, partial
(63%)
Length = 1165
Score = 207 bits (526), Expect = 1e-53
Identities = 119/343 (34%), Positives = 181/343 (52%), Gaps = 10/343 (2%)
Frame = +3
Query: 256 CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNT 307
CL + YL+ I + +A+ + V W +A LLT +G KH C+ +
Sbjct: 9 CL*SSLQYLKNFQKRQVPILERFALLITTTVIWAYAHLLTASGAYKHRPDVTQHSCRTDR 188
Query: 308 SDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTP 367
++ ++S PW + PYPL+WG P F+ + M L+S V+S G + +S LA++ PP
Sbjct: 189 ANLISSAPWIKIPYPLEWGAPTFDAGHSFGMMAAVLVSLVESTGAFKAASRLASATPPPA 368
Query: 368 GVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLF 427
VLSRGIG +G LL GL+GT GST ENV + ++GSRR +Q+ A +I ++
Sbjct: 369 HVLSRGIGWQGIGILLNGLFGTLTGSTVSVENVGLLGSNRVGSRRVIQVSAGFMIFFAML 548
Query: 428 GKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPA 487
GK G ASIP + A + C+++ ++ ++GLS L++T S RN+ I G++LF LSIP
Sbjct: 549 GKFGALFASIPFPIFAAIYCVLFGLVASVGLSFLQFTNMNSMRNLFITGVALFLGLSIPE 728
Query: 488 YFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILD 547
YF++Y I HGP +K N LN IF +A ++A+ LD
Sbjct: 729 YFREYT----------------IRALHGPAHTKAGWFNDFLNTIFYSSPTVALIIAVFLD 860
Query: 548 NTV--PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
NT+ S ++R + W+K + D Y LP + R F
Sbjct: 861 NTLDYKDSAKDRGMPWWAKFRTFKADSRNEEFYSLPFNLNRFF 989
>AW697093 similar to GP|20466159|gb putative membrane transporter
{Arabidopsis thaliana}, partial (18%)
Length = 603
Score = 114 bits (284), Expect(2) = 9e-52
Identities = 58/70 (82%), Positives = 62/70 (87%), Gaps = 1/70 (1%)
Frame = +2
Query: 155 IFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNEN-KFKHIMKELQGAIIIGSAFQTLLG 213
+ G RLPLIQGPSFVYLAP LAIINSPE Q LN N KFKHIM+ELQGAIIIGSAFQ LLG
Sbjct: 383 VLGQRLPLIQGPSFVYLAPALAIINSPELQGLNGNDKFKHIMRELQGAIIIGSAFQALLG 562
Query: 214 YTGLMSLLVR 223
YTGLMSLL++
Sbjct: 563 YTGLMSLLIK 592
Score = 108 bits (270), Expect(2) = 9e-52
Identities = 60/112 (53%), Positives = 74/112 (65%), Gaps = 3/112 (2%)
Frame = +1
Query: 52 SPSDGVPTNNARVEERTTRLPVMVDH---DDLVLRRRPSPLNYELTDSPALVFLAVYGIQ 108
+P+ G T A RT R +VD DD R + + YEL D P LV +AVYGIQ
Sbjct: 64 NPAPGTGTEQATQVRRTLRNEEVVDGLVVDDEGFASRHAHMKYELRDFPGLVPIAVYGIQ 243
Query: 109 HYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHTIFGSRL 160
HY+S++GSLIL PLVI PAMG SH+ET+ +V TVL VSG+TTLLH FGS +
Sbjct: 244 HYVSMLGSLILIPLVIVPAMGGSHEETSNVVSTVLFVSGLTTLLHISFGSEI 399
>CA990413 similar to PIR|T05632|T05 hypothetical protein F20D10.170 -
Arabidopsis thaliana, partial (22%)
Length = 731
Score = 199 bits (506), Expect = 2e-51
Identities = 95/189 (50%), Positives = 132/189 (69%)
Frame = +1
Query: 407 KMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTET 466
K+ SRR ++LGA LI+ S GKVG +ASIP+A+ A +LC MWA+ ALGLS L+Y ++
Sbjct: 1 KVASRRVLELGAVFLILFSFVGKVGALLASIPQALAAAILCFMWALTVALGLSTLQYGQS 180
Query: 467 GSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNY 526
S RN+ IVG++LF +SIP+YFQQY+ PES+ +PSY PY S GPF S ++L++
Sbjct: 181 PSFRNMTIVGVALFLGMSIPSYFQQYQ--PESSLILPSYLVPYAAASSGPFHSGLKQLDF 354
Query: 527 VLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGR 586
+N + S++MV+ LVA +LDNTVPGSKQER +Y WS+ D D + SEY LP ++
Sbjct: 355 AINALMSMNMVVTLLVAFLLDNTVPGSKQERGVYTWSRAEDIAADASLQSEYSLPKKLAW 534
Query: 587 CFRWVKWVG 595
C W+K +G
Sbjct: 535 CCCWLKCLG 561
>AW689217 similar to GP|15144504|gb| putative permease {Lycopersicon
esculentum}, partial (16%)
Length = 609
Score = 100 bits (249), Expect(3) = 2e-27
Identities = 44/81 (54%), Positives = 64/81 (78%)
Frame = +2
Query: 85 RPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLL 144
RPS + + LT++P LV L Y +QHYLS+IGSL+L PLV+ P MG + ++TA ++ T+L
Sbjct: 257 RPSDMKFGLTENPGLVPLVYYSLQHYLSLIGSLVLIPLVMVPTMGGTDNDTANVISTMLF 436
Query: 145 VSGVTTLLHTIFGSRLPLIQG 165
+SG+TT+LH+ FG+RLPL+QG
Sbjct: 437 LSGITTILHSYFGTRLPLVQG 499
Score = 30.8 bits (68), Expect(3) = 2e-27
Identities = 12/19 (63%), Positives = 16/19 (84%)
Frame = +1
Query: 183 FQELNENKFKHIMKELQGA 201
F+ L +KF+HIM+ELQGA
Sbjct: 553 FRNLTHHKFRHIMRELQGA 609
Score = 29.6 bits (65), Expect(3) = 2e-27
Identities = 13/17 (76%), Positives = 14/17 (81%)
Frame = +3
Query: 167 SFVYLAPVLAIINSPEF 183
SFVYLAP L I+NS EF
Sbjct: 504 SFVYLAPALVIMNSEEF 554
>BM815623 similar to GP|14334908|gb putative membrane transporter protein
{Arabidopsis thaliana}, partial (29%)
Length = 461
Score = 118 bits (296), Expect = 6e-27
Identities = 63/166 (37%), Positives = 94/166 (55%)
Frame = +1
Query: 385 GLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAG 444
GL+GT G + ENV + T++GSRR +Q+ A +I S+ GK G F ASIP + A
Sbjct: 1 GLFGTLTGPSVSVENVGLLGSTRVGSRRVIQISAGFMIFFSMLGKFGAFFASIPFPIFAA 180
Query: 445 LLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPS 504
+ C+++ ++ ++GLS L++T S R++ I G+SLF LSIP YF+++ S
Sbjct: 181 MYCVLFGLVASVGLSFLQFTNMNSMRSLFITGVSLFLGLSIPEYFREFTSK--------- 333
Query: 505 YFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV 550
HGP +K N LN IF +AF+VA+ LDNT+
Sbjct: 334 -------ALHGPAHTKARWFNDFLNTIFFSSSTVAFIVAVFLDNTL 450
>TC93725 similar to GP|10177467|dbj|BAB10858. permease 1 {Arabidopsis
thaliana}, partial (48%)
Length = 985
Score = 69.3 bits (168), Expect(2) = 9e-24
Identities = 30/98 (30%), Positives = 58/98 (58%)
Frame = +3
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
+++ +T P + G QHYL ++G+ +L P + P MG ++E A ++ T+L V+G+
Sbjct: 267 VSFCITSPPPWPEAILLGFQHYLVMLGTTVLIPSSLVPQMGGGNEEKAKVIQTLLFVAGI 446
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQEL 186
T T FG+RLP + G S+ ++ ++II + + ++
Sbjct: 447 NTFFQTTFGTRLPAVIGGSYTFVPTTISIILAGRYSDI 560
Score = 59.3 bits (142), Expect(2) = 9e-24
Identities = 39/126 (30%), Positives = 62/126 (48%), Gaps = 6/126 (4%)
Frame = +1
Query: 191 FKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LA 244
F+ IM+ QGA+I+ S Q +LG++GL +VRF++P+ +A G F LA
Sbjct: 580 FEKIMRGTQGALIVASTLQIVLGFSGLWRNVVRFLSPLSAVPLVALSGFGLYEFGFPVLA 759
Query: 245 RVLRSVQYRY*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQ 304
+ + L+ F Y+ + IF +AV +A+ W +A+LLT G K+
Sbjct: 760 KCVEIGLPEIIILVVFSQYIPHMMKGEKPIFDRFAVIFSVAIVWLYAYLLTVGGAYKNSA 939
Query: 305 VNTSDT 310
T T
Sbjct: 940 PKTQIT 957
>BG452976 weakly similar to GP|15983805|gb At1g49960/F2J10_14 {Arabidopsis
thaliana}, partial (33%)
Length = 610
Score = 98.2 bits (243), Expect = 8e-21
Identities = 53/149 (35%), Positives = 88/149 (58%), Gaps = 4/149 (2%)
Frame = +2
Query: 97 PALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHTIF 156
P +FL G QHYL ++G++++ ++ P MG + E A M+ T+L V+G+ TLL T
Sbjct: 110 PEGIFL---GFQHYLVMLGTIVIVSTILVPLMGGGNVEKAEMIQTLLFVAGINTLLQTWL 280
Query: 157 GSRLPLIQGPSFVYLAPVLAIINSPEFQE-LN-ENKFKHIMKELQGAIIIGSAFQTLLGY 214
G+RLP++ G SF ++ P +++ S LN +FK M+ +QGA++I S Q ++G+
Sbjct: 281 GTRLPVVIGASFAFIIPAISVAFSSRMSVFLNPRQRFKQSMRAIQGALLIASFVQGMIGF 460
Query: 215 TGLMSLLVRFINPV--VVSSTIAAVGLFL 241
G + RF P+ V T+ +GLF+
Sbjct: 461 FGFWGIFGRFFXPLSAVPLVTLTGLGLFV 547
>TC89886 weakly similar to GP|14587294|dbj|BAB61205. putative permease 1
{Oryza sativa (japonica cultivar-group)}, partial (25%)
Length = 638
Score = 82.4 bits (202), Expect = 4e-16
Identities = 54/170 (31%), Positives = 84/170 (48%), Gaps = 6/170 (3%)
Frame = +1
Query: 421 LIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLF 480
+I+ S+FGK G F ASIP + A + +++ ++ A +S +++ S RNI + GL+LF
Sbjct: 10 MILCSIFGKFGAFFASIPLPIFAAIY*VLFGIVAATRISFIQFANNNSIRNIYVFGLTLF 189
Query: 481 FSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAF 540
+SIP YF ++P+ HGP R+ N +LN IFS +A
Sbjct: 190 LGISIPQYFVM-NTAPD---------------GHGPVRTNGGWFNDILNTIFSSPPTVAI 321
Query: 541 LVALILDNTVPGSKQ--ERELYGW----SKPNDAREDPFIVSEYGLPARV 584
+V +LDNT+ + +R L W + D R D F Y P R+
Sbjct: 322 IVGTVLDNTLEAKQTAVDRGLPWWVPFQKRKGDVRNDEF----YRFPLRL 459
>BQ149045 similar to PIR|T02307|T02 probable membrane transporter At2g34190
[imported] - Arabidopsis thaliana, partial (38%)
Length = 650
Score = 79.3 bits (194), Expect = 4e-15
Identities = 56/201 (27%), Positives = 97/201 (47%), Gaps = 16/201 (7%)
Frame = +2
Query: 176 AIINSPEFQELNEN--KFKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSST 233
AII+ + + + +F M+ +QGA+I S+ Q +LG++ L ++ R +P+ +
Sbjct: 14 AIIHDSKLASIEDPHLRFVKTMRAVQGAMIAASSIQIILGFSQLWAICSRXFSPLGMVPV 193
Query: 234 IAAVGLFL*---LARVLRSVQYRY*CLLFFVL---YLRKISVFGHHIFQIYAVPLGLAVT 287
IA G L V V+ L+ V YL+ + I + +A+ + +
Sbjct: 194 IALSGFGLFNRGFPVVGHCVETGIPMLILXVAFSQYLKNFNARQLPILERFALLISTTII 373
Query: 288 WTFAFLLTENGRMK--------HCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMC 339
W +A LLT +G K +C+ + S+ ++S PW + PYP + G P F+ A M
Sbjct: 374 WAYAHLLTTSGTYKLRSELTQYNCRTDKSNLISSXPWMKIPYPFEXGFPTFDXGHAFGMM 553
Query: 340 VVSLISSVDSVGTYHTSSLLA 360
L+S ++S G Y +S LA
Sbjct: 554 XAVLVSLIESTGAYTAASXLA 616
>BF637188 similar to GP|14334908|gb putative membrane transporter protein
{Arabidopsis thaliana}, partial (19%)
Length = 448
Score = 73.2 bits (178), Expect = 3e-13
Identities = 31/85 (36%), Positives = 53/85 (61%)
Frame = +2
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P+ V + G QHY+ +G+ ++ P + P+MG + D+ +V T+L V G+
Sbjct: 188 LEYCIDSNPSWVETILLGFQHYILALGTAVMIPSFLVPSMGGNDDDKVRVVQTLLFVEGI 367
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAP 173
TLL T+FG+RLP + G S+ ++ P
Sbjct: 368 NTLLQTLFGTRLPTVIGGSYAFMCP 442
>AW683939 similar to GP|20466159|gb| putative membrane transporter
{Arabidopsis thaliana}, partial (9%)
Length = 309
Score = 65.1 bits (157), Expect = 7e-11
Identities = 30/56 (53%), Positives = 37/56 (65%)
Frame = +2
Query: 540 FLVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCFRWVKWVG 595
FL+ L VPGS+QER +Y WS+ AR +P + +Y LP R GR FRWVKWVG
Sbjct: 44 FLLLLSWIILVPGSRQERGVYVWSEAEVARREPAVAKDYELPWRAGRIFRWVKWVG 211
Score = 36.6 bits (83), Expect = 0.028
Identities = 17/22 (77%), Positives = 20/22 (90%)
Frame = +1
Query: 528 LNMIFSLHMVIAFLVALILDNT 549
+N + SLHMVIAFLVA+ILDNT
Sbjct: 7 VNTLCSLHMVIAFLVAVILDNT 72
>AW695592 weakly similar to PIR|T00984|T009 probable membrane transporter
At2g26510 [imported] - Arabidopsis thaliana, partial
(10%)
Length = 173
Score = 32.7 bits (73), Expect = 0.40
Identities = 16/53 (30%), Positives = 25/53 (46%), Gaps = 8/53 (15%)
Frame = +1
Query: 278 YAVPLGLAVTWTFAFLLTENGRMK--------HCQVNTSDTMTSPPWFRFPYP 322
+A+ + +A+ W FA +LT G C+ + S +T PW PYP
Sbjct: 10 FALLICIAIIWAFAAILTVAGAYNTSKEKTQTSCRTDRSYLLTRAPWIYVPYP 168
>AL373498 similar to PIR|D84651|D84 hypothetical protein At2g25680 [imported]
- Arabidopsis thaliana, partial (30%)
Length = 463
Score = 30.8 bits (68), Expect = 1.5
Identities = 33/107 (30%), Positives = 49/107 (44%), Gaps = 1/107 (0%)
Frame = +2
Query: 141 TVLLVSGVTTLLH-TIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQ 199
T L+ +GV +L I+G +P+ P+ +I + L++ F + E+
Sbjct: 14 TTLIFTGVCNMLTGVIYGVPMPV---------QPMKSI----SAEALSDTTFG--VPEIM 148
Query: 200 GAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARV 246
A I+ S LLG TGLM L+ +FI VV A GL L V
Sbjct: 149 AAGIMTSGAVLLLGVTGLMQLVYKFIPLSVVRGIQLAQGLSFALTAV 289
>AW773872 similar to GP|21595706|gb unknown {Arabidopsis thaliana}, partial
(25%)
Length = 671
Score = 30.4 bits (67), Expect = 2.0
Identities = 20/97 (20%), Positives = 44/97 (44%), Gaps = 6/97 (6%)
Frame = +3
Query: 14 ETKAGQSLSRQPDLEASPVVSTPSQAVNDVPHGDKVPPSPSDGVPTNNARVEERTTRLPV 73
E + S +P ++ + + S PSQ ++ P+ + + P+PS P++ + +T P
Sbjct: 69 EAEDDDSEFEEPSIDPASLRSPPSQFLSTDPNPNPINPTPSPSPPSDLPKSTPPSTTDPQ 248
Query: 74 MVDH------DDLVLRRRPSPLNYELTDSPALVFLAV 104
D+ D+ ++ +P E+ L+ A+
Sbjct: 249 STDNTNTTASDNQNVKPQPRSFTVEIVCGSFLIMFAL 359
>BF639866 homologue to GP|3036792|emb| putative protein (fragment)
{Arabidopsis thaliana}, partial (3%)
Length = 427
Score = 30.0 bits (66), Expect = 2.6
Identities = 15/30 (50%), Positives = 19/30 (63%), Gaps = 1/30 (3%)
Frame = +1
Query: 554 KQERELY-GWSKPNDAREDPFIVSEYGLPA 582
K+ERELY WS PN+ R P + GLP+
Sbjct: 280 KKERELYLMWSSPNEIRTTPS*PNNVGLPS 369
>TC82424 similar to PIR|T51283|T51283 glucan 1 3-beta-glucosidase (EC
3.2.1.58) [imported] - common tobacco, partial (45%)
Length = 962
Score = 29.6 bits (65), Expect = 3.4
Identities = 27/82 (32%), Positives = 40/82 (47%), Gaps = 4/82 (4%)
Frame = -3
Query: 252 YRY*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNTSDTM 311
+R CLLF +L + +S+ HH+FQ + V L L+ L +N + H + D+
Sbjct: 600 HRIHCLLFHILVQKLLSLSCHHLFQ-HMVSLQLSQ------LHVKNHWILHSLM--KDSP 448
Query: 312 TSPPWF----RFPYPLQWGTPV 329
PWF RF L+ G PV
Sbjct: 447 RRIPWFLCRLRFL*ALRIGVPV 382
>CB891067 similar to PIR|C86334|C863 hypothetical protein AAF79906.1
[imported] - Arabidopsis thaliana, partial (8%)
Length = 538
Score = 28.9 bits (63), Expect = 5.8
Identities = 21/76 (27%), Positives = 35/76 (45%), Gaps = 11/76 (14%)
Frame = -1
Query: 451 AMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSI-----------PAYFQQYESSPESN 499
A L L +S +ETGSSR+ +++GL+ F + PA + E P +
Sbjct: 508 AELAILRISISGKSETGSSRDTVLLGLTENFGIGKELGGGEFE*VGPAQSSESEKLPLGS 329
Query: 500 FSVPSYFQPYIVTSHG 515
+ S+ QP + + G
Sbjct: 328 TKILSFLQPLSLIATG 281
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,100,072
Number of Sequences: 36976
Number of extensions: 291960
Number of successful extensions: 1926
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 1883
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1903
length of query: 595
length of database: 9,014,727
effective HSP length: 102
effective length of query: 493
effective length of database: 5,243,175
effective search space: 2584885275
effective search space used: 2584885275
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148969.3


