

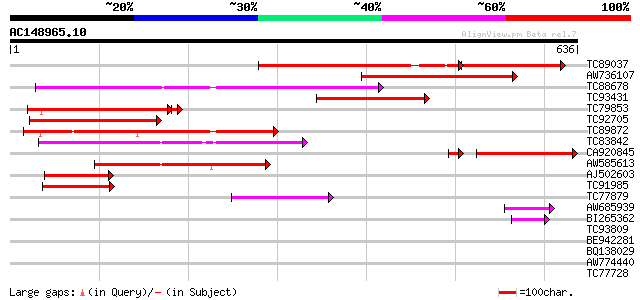
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148965.10 + phase: 0
(636 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89037 similar to GP|13605811|gb|AAK32891.1 AT5g36940/MLF18_60 ... 238 8e-97
AW736107 similar to GP|13605811|gb| AT5g36940/MLF18_60 {Arabidop... 333 2e-91
TC88678 similar to PIR|S51171|S51171 amino acid transport protei... 258 6e-69
TC93431 similar to GP|13605811|gb|AAK32891.1 AT5g36940/MLF18_60 ... 242 3e-64
TC79853 similar to GP|13605811|gb|AAK32891.1 AT5g36940/MLF18_60 ... 220 2e-57
TC92705 similar to GP|13605811|gb|AAK32891.1 AT5g36940/MLF18_60 ... 219 2e-57
TC89872 similar to GP|14140148|emb|CAC39065. putative protein {O... 216 2e-56
TC83842 similar to PIR|S51171|S51171 amino acid transport protei... 200 1e-51
CA920845 weakly similar to GP|10178213|dbj cationic amino acid t... 149 2e-38
AW585613 PIR|T48473|T48 amino acid transport-like protein - Arab... 139 3e-33
AJ502603 weakly similar to GP|2116759|dbj| YfnA {Bacillus subtil... 77 2e-14
TC91985 similar to PIR|A86307|A86307 amino acid transporter homo... 64 2e-10
TC77879 similar to PIR|D86442|D86442 probable amino acid permeas... 49 6e-06
AW685939 similar to GP|13430818|gb| unknown protein {Arabidopsis... 47 2e-05
BI265362 weakly similar to GP|7296598|gb|A CG11128 gene product ... 43 4e-04
TC93809 similar to PIR|H86428|H86428 hypothetical protein AAF197... 40 0.003
BE942281 similar to GP|8567793|gb| amino acid transporter putat... 39 0.008
BQ138029 weakly similar to GP|1764098|gb|A putative permease {Ur... 37 0.018
AW774440 weakly similar to PIR|T48473|T484 amino acid transport-... 30 2.2
TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance... 30 3.7
>TC89037 similar to GP|13605811|gb|AAK32891.1 AT5g36940/MLF18_60
{Arabidopsis thaliana}, partial (39%)
Length = 1337
Score = 238 bits (606), Expect(2) = 8e-97
Identities = 121/231 (52%), Positives = 169/231 (72%), Gaps = 1/231 (0%)
Frame = +3
Query: 280 ALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLL 339
+L++CC LYM+VS V+VGLVPYY++NP+TPISSAF+ GM+WA YII GA+TAL SSLL
Sbjct: 6 SLSLCCGLYMIVSIVVVGLVPYYDINPNTPISSAFAVNGMQWAAYIINAGALTALCSSLL 185
Query: 340 GSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMV 399
G++LPQPR+ MAMARDGLLP FFSDI++ + +P+KSTIVTGL A LAFFMDVSQL+G+V
Sbjct: 186 GTMLPQPRILMAMARDGLLPPFFSDINKHSHVPVKSTIVTGLVPAALAFFMDVSQLSGLV 365
Query: 400 SVGTLLAFTTVAVSVLIIRYVPPDEIPIPASLLTSVD-PLLRHSGDDIEEDRTVSPVDLA 458
SVGTL+AFT A++VLI+RY+PP E+P+P S +D + +S +E ++
Sbjct: 366 SVGTLIAFTISAITVLIVRYIPPIEVPLPHSRQEPIDSESMEYSWSHLETNK-------- 521
Query: 459 SYSDNSHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGIL 509
+ ++ L K D ++PLI K + +++N R+K+ IA +C+G +
Sbjct: 522 NDANRKPLIVKEDASTDYPLIAKYLAIDKYN-GNRKKIVGCVIASICLGCI 671
Score = 135 bits (339), Expect(2) = 8e-97
Identities = 63/117 (53%), Positives = 82/117 (69%)
Frame = +1
Query: 507 GILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFV 566
G+ +++ +A + +R TL G G ++ L + L CI QDD RH FG +GGF CP V
Sbjct: 664 GVFVLTFAACSAYLLSSVRFTLCGVGGILLLSGFVFLTCIDQDDERHNFGSTGGFKCPLV 843
Query: 567 PFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHSSLLNAIYVPSAR 623
P LP CILIN+YLLI G TWLRVSVWL IGV+IY+FYGRTHSSL +A++V +A+
Sbjct: 844 PLLPITCILINSYLLISRGGATWLRVSVWLAIGVIIYVFYGRTHSSLRDAVHVKAAQ 1014
>AW736107 similar to GP|13605811|gb| AT5g36940/MLF18_60 {Arabidopsis
thaliana}, partial (9%)
Length = 578
Score = 333 bits (853), Expect = 2e-91
Identities = 169/175 (96%), Positives = 169/175 (96%)
Frame = +3
Query: 395 LAGMVSVGTLLAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSP 454
L V V TLLAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSP
Sbjct: 54 LGMQVGVLTLLAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSP 233
Query: 455 VDLASYSDNSHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGS 514
VDLASYSDNSHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGS
Sbjct: 234 VDLASYSDNSHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGS 413
Query: 515 ASAERCPRILRVTLFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFL 569
ASAERCPRILRVTLFGAGVVIFLCSIIVLACIKQDDTRHTFGHS GFACPFVPFL
Sbjct: 414 ASAERCPRILRVTLFGAGVVIFLCSIIVLACIKQDDTRHTFGHSRGFACPFVPFL 578
>TC88678 similar to PIR|S51171|S51171 amino acid transport protein AAT1 -
Arabidopsis thaliana, partial (72%)
Length = 1520
Score = 258 bits (658), Expect = 6e-69
Identities = 141/390 (36%), Positives = 232/390 (59%)
Frame = +3
Query: 30 IHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAAL 89
I R ++ + L+ DL+ G+GA +G+G+++L G A++ AGPA+V+S I+GI+A L
Sbjct: 222 IKARSGNEMKKTLNWWDLMWFGIGAVVGSGIFVLTGLEAKQHAGPAVVLSFVISGISALL 401
Query: 90 SALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALF 149
S CY E A P AG ++ Y + +G+ VA++ +++LEY IG +AVAR T A
Sbjct: 402 SVFCYTEFAVEIPVAGGSFAYLRVEMGDFVAFIAAGNILLEYVIGNAAVARSWTSYFATL 581
Query: 150 FGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVSVM 209
+ + + P G + DP A +V IT L K SS I T +++V+
Sbjct: 582 CNKNPDDFRIIVHNMNPDYGHL-DPIAIGALVAITALAVYSTKGSSIFNYIATMFHMAVI 758
Query: 210 LFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNP 269
+FI+I G + + + P+G++GM + SA++FF+YIGFD+V++ AEE KNP
Sbjct: 759 IFIVIAGLIKAKPENF------NDFTPFGLHGMVSSSAVLFFAYIGFDAVSTMAEETKNP 920
Query: 270 QRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTG 329
RD+PIG+ ++ I +Y L+ A + + Y EL+ D P S AFS+ GM+WA YI++ G
Sbjct: 921 GRDIPIGLVGSMTITTAIYCLLGATLCLMQNYKELDTDAPFSVAFSAVGMDWAKYIVSLG 1100
Query: 330 AVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFF 389
A+ + + LL S + Q R +AR ++P +F+ + RT P+ +TI + A++AFF
Sbjct: 1101ALKGMTTVLLVSAVGQARYLTHIARTHMMPPWFALVDERTGTPMNATISMLIATAIVAFF 1280
Query: 390 MDVSQLAGMVSVGTLLAFTTVAVSVLIIRY 419
++S L+ ++S+ TL F+ VA+++L+ RY
Sbjct: 1281TNLSILSSLLSISTLFIFSLVALALLVRRY 1370
>TC93431 similar to GP|13605811|gb|AAK32891.1 AT5g36940/MLF18_60
{Arabidopsis thaliana}, partial (12%)
Length = 382
Score = 242 bits (618), Expect = 3e-64
Identities = 124/126 (98%), Positives = 125/126 (98%)
Frame = +2
Query: 345 QPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTL 404
+ RVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTL
Sbjct: 5 EARVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTL 184
Query: 405 LAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNS 464
LAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNS
Sbjct: 185 LAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNS 364
Query: 465 HLHDKS 470
HLHDKS
Sbjct: 365 HLHDKS 382
>TC79853 similar to GP|13605811|gb|AAK32891.1 AT5g36940/MLF18_60
{Arabidopsis thaliana}, partial (23%)
Length = 782
Score = 220 bits (560), Expect(2) = 2e-57
Identities = 109/168 (64%), Positives = 130/168 (76%), Gaps = 6/168 (3%)
Frame = +3
Query: 21 LIRRKQVDSIHYRG------HPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGP 74
L RRK+VD+ H QLA++L+V DL+ IGVGATIGAGVY+L+GTVARE AGP
Sbjct: 15 LTRRKKVDNSHVENINKNSHGVQLAKELTVPDLMAIGVGATIGAGVYVLVGTVAREHAGP 194
Query: 75 ALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIG 134
AL +S +AGIAAALSALCYAELA R PSAGSAYHY YIC+GEGVAWL+GWSL LE IG
Sbjct: 195 ALPLSFMVAGIAAALSALCYAELASRFPSAGSAYHYAYICLGEGVAWLIGWSLTLEVVIG 374
Query: 135 ASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVL 182
+ +ARGITPNLA GG DNLP FL+ +PG+ IVVDP AA+++ +
Sbjct: 375 GAVIARGITPNLAALIGGMDNLPGFLSYQHIPGIDIVVDPLAAIVVFI 518
Score = 21.6 bits (44), Expect(2) = 2e-57
Identities = 8/12 (66%), Positives = 10/12 (82%)
Frame = +1
Query: 182 LITLLLCLGIKE 193
L+T +LC GIKE
Sbjct: 517 LVTWILCTGIKE 552
>TC92705 similar to GP|13605811|gb|AAK32891.1 AT5g36940/MLF18_60
{Arabidopsis thaliana}, partial (22%)
Length = 698
Score = 219 bits (559), Expect = 2e-57
Identities = 105/148 (70%), Positives = 123/148 (82%)
Frame = +3
Query: 23 RRKQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFI 82
RRKQVDS P LA++LSV L+ IGVG+TIGAGVY+L+GTVARE +GPAL IS I
Sbjct: 255 RRKQVDSPRQNTKPLLAKELSVYHLIAIGVGSTIGAGVYVLVGTVAREHSGPALAISFLI 434
Query: 83 AGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGI 142
AG+AA LSA CYAELACRCPSAGSAYHY+YIC+GE VAWL+GW+L+LEYTIG++AVARGI
Sbjct: 435 AGLAAGLSAFCYAELACRCPSAGSAYHYSYICLGESVAWLIGWALLLEYTIGSAAVARGI 614
Query: 143 TPNLALFFGGQDNLPSFLARHTLPGLGI 170
TPNLA FGG DNL FL+R +PG+ I
Sbjct: 615 TPNLAPLFGGLDNLTVFLSRQHIPGIDI 698
>TC89872 similar to GP|14140148|emb|CAC39065. putative protein {Oryza
sativa}, partial (43%)
Length = 927
Score = 216 bits (550), Expect = 2e-56
Identities = 121/297 (40%), Positives = 184/297 (61%), Gaps = 11/297 (3%)
Frame = +1
Query: 16 RGFGSLIRRKQVDSIHYR------GHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAR 69
R S +R K+V S + GH L+R+L V DL+ +G+GA+IGAG++++ GTVA
Sbjct: 52 RFLSSALRSKRVISPSEKTARDNSGHG-LSRRLGVFDLILLGIGASIGAGIFVVTGTVAH 228
Query: 70 EQAGPALVISLFIAGIAAALSALCYAELACRCPSA-GSAYHYTYICIGEGVAWLVGWSLI 128
+ GPA+ +S AG + ++ALCYAELA R P+ G AY YTY E A+LV L+
Sbjct: 229 D-VGPAVTLSFIFAGASCVINALCYAELASRFPAVVGGAYLYTYTAFNELTAFLVFSQLM 405
Query: 129 LEYTIGASAVARG----ITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLIT 184
L+Y IGA+++AR I L +F +DN+P ++ G + ++ A +L++L+T
Sbjct: 406 LDYHIGAASIARSLANYIVTFLEIFPVFKDNIPKWIGHGLNIGNVLSINALAPILLILLT 585
Query: 185 LLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFA 244
L+LCLG++ESS V S +T + +++ +I G + + W S + P G+ +F
Sbjct: 586 LILCLGVRESSAVNSFMTVTKIIIVIVVIFAGAFEVDVSNW------SPFVPNGIRPVFT 747
Query: 245 GSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPY 301
G+ +VFF+Y+GFD+V ++AEE K PQRDLPIGI +L +C LY+ V VI G+VPY
Sbjct: 748 GATVVFFAYVGFDAVANSAEESKKPQRDLPIGIIGSLLVCIALYIGVCLVITGMVPY 918
>TC83842 similar to PIR|S51171|S51171 amino acid transport protein AAT1 -
Arabidopsis thaliana, partial (55%)
Length = 1295
Score = 200 bits (509), Expect = 1e-51
Identities = 112/302 (37%), Positives = 173/302 (57%)
Frame = +3
Query: 33 RGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSAL 92
R Q+ + L+ DL+ G+GA +G+G+++L G ARE+AGPA+V+S ++GI+A LS
Sbjct: 411 RSSHQMKKTLNGWDLIWFGIGAVVGSGIFVLTGLEAREEAGPAVVLSYAVSGISALLSVF 590
Query: 93 CYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGG 152
CY E A P AG ++ Y + +G+ VA++ +++LEY IGA+AVAR T A
Sbjct: 591 CYTEFAVEIPVAGGSFAYLRVELGDFVAFIAAGNILLEYVIGAAAVARSWTSYFATLCNK 770
Query: 153 QDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVSVMLFI 212
N + + P G +DP A + ++ I L L + SS I T ++ V+ FI
Sbjct: 771 NPNDFRIIFHNMNPDYG-HLDPIAVIALIAIATLAVLSTRISSLFNKIATIFHLVVIAFI 947
Query: 213 IIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRD 272
IIVG A Y + + P+G G+F SA++FF+YIGFD+V++ AEE KNP RD
Sbjct: 948 IIVG---LINANPENY---ASFAPFGTRGVFKASAVLFFAYIGFDAVSTMAEETKNPGRD 1109
Query: 273 LPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVT 332
+PIG+ ++ I +Y ++ + + Y ++ + P S AFSS G WA YI GA+
Sbjct: 1110IPIGLVGSMVIITFIYCSLATTLCLMQNYKTIDVNAPFSVAFSSVGWGWAKYIGALGALK 1289
Query: 333 AL 334
+
Sbjct: 1290GM 1295
>CA920845 weakly similar to GP|10178213|dbj cationic amino acid
transporter-like protein {Arabidopsis thaliana}, partial
(14%)
Length = 697
Score = 149 bits (377), Expect(2) = 2e-38
Identities = 68/113 (60%), Positives = 84/113 (74%)
Frame = -1
Query: 524 LRVTLFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLID 583
+R L G G + + + L C+ QDD RH FGHSGGF CPFVP LP ACILIN+YLLI+
Sbjct: 598 VRFALCGVGGTLLVSGFVFLTCMDQDDARHDFGHSGGFVCPFVPLLPVACILINSYLLIN 419
Query: 584 LGVDTWLRVSVWLLIGVLIYLFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
LG +TWLRVSVWL G+LIY FYGRTHSSL +AIYVP+++ DE ++ +H A
Sbjct: 418 LGAETWLRVSVWLATGLLIYGFYGRTHSSLKDAIYVPASQVDERYQPPRSHAA 260
Score = 28.1 bits (61), Expect(2) = 2e-38
Identities = 9/17 (52%), Positives = 14/17 (81%)
Frame = -2
Query: 493 RRKLAAWTIALLCIGIL 509
RR++ WTIA++C+G L
Sbjct: 690 RRRVVGWTIAIICLGAL 640
>AW585613 PIR|T48473|T48 amino acid transport-like protein - Arabidopsis
thaliana, partial (34%)
Length = 603
Score = 139 bits (350), Expect = 3e-33
Identities = 72/200 (36%), Positives = 121/200 (60%), Gaps = 3/200 (1%)
Frame = +2
Query: 96 ELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDN 155
+ A P AG A+ Y I GE A+ G +L+++Y + +AV+R T L FG +
Sbjct: 5 QFAVHLPVAGGAFSYIRITFGEFPAFFTGANLVMDYVMSNAAVSRSFTAYLGTAFGISTS 184
Query: 156 LPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVSVMLFIIIV 215
F+ G +DP A +++++IT+++C +ESS V I+T +++ + F+I++
Sbjct: 185 KWRFVVSGLPKGFN-EIDPVAVLVVLVITVIICCSTRESSKVNMIMTAFHIAFIFFVIVM 361
Query: 216 GGYLGFKAGW---VGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRD 272
G G E PSG+FP+G G+F G+A+V+ SYIG+D+V++ AEEV+NP +D
Sbjct: 362 GFIKGDSKNLSSPANPEHPSGFFPFGAAGVFNGAAMVYLSYIGYDAVSTMAEEVENPVKD 541
Query: 273 LPIGISTALAICCVLYMLVS 292
+P+G+S ++AI VLY L++
Sbjct: 542 IPVGVSGSVAIVTVLYCLMA 601
>AJ502603 weakly similar to GP|2116759|dbj| YfnA {Bacillus subtilis}, partial
(16%)
Length = 450
Score = 77.4 bits (189), Expect = 2e-14
Identities = 37/77 (48%), Positives = 52/77 (67%)
Frame = +1
Query: 40 RKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAELAC 99
R LS DL+ IG+G IG G+ ++ G A +AGPA+VIS I+GIAA+ +AL Y+EL+
Sbjct: 220 RTLSAFDLIMIGLGGIIGTGILVITGQAAATKAGPAVVISFIISGIAASFAALSYSELSS 399
Query: 100 RCPSAGSAYHYTYICIG 116
P +GSAY + Y +G
Sbjct: 400 MIPISGSAYTFVYATLG 450
>TC91985 similar to PIR|A86307|A86307 amino acid transporter homolog
[imported] - Arabidopsis thaliana, partial (29%)
Length = 661
Score = 63.5 bits (153), Expect = 2e-10
Identities = 30/80 (37%), Positives = 50/80 (62%)
Frame = +1
Query: 38 LARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAEL 97
+ R L+ DL + GA +G+G++++ G AR AGP++++S +G +A LSAL YAE
Sbjct: 283 MTRCLTWWDLTWLAFGAVVGSGIFVVTGQEARFDAGPSIILSYAASGFSALLSALIYAEF 462
Query: 98 ACRCPSAGSAYHYTYICIGE 117
A P AG ++ + I +G+
Sbjct: 463 AVEVPVAGGSFSFLRIELGD 522
>TC77879 similar to PIR|D86442|D86442 probable amino acid permease [imported]
- Arabidopsis thaliana, partial (81%)
Length = 2037
Score = 48.9 bits (115), Expect = 6e-06
Identities = 32/116 (27%), Positives = 54/116 (45%), Gaps = 1/116 (0%)
Frame = +2
Query: 249 VFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYY-ELNPD 307
+F++ +DS+++ A EV+NP+++LP G+ AL + V Y + G VP EL D
Sbjct: 893 LFWNLNYWDSISTLAGEVENPKKNLPKGLFYALILVVVAYFFPLLIGTGAVPVQRELWTD 1072
Query: 308 TPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFS 363
S G W + + A + + + + MA G+LP FF+
Sbjct: 1073 GYFSEIAMIIGGVWLRWWLQAAAAMSNMGMFVAEMSSDSYQLLGMAERGMLPEFFT 1240
>AW685939 similar to GP|13430818|gb| unknown protein {Arabidopsis thaliana},
partial (9%)
Length = 584
Score = 47.0 bits (110), Expect = 2e-05
Identities = 21/56 (37%), Positives = 30/56 (53%)
Frame = -3
Query: 556 GHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHS 611
G S GF CP VP +P CI N +L L + W+R + + V +Y YG+ H+
Sbjct: 468 GFS*GFLCPGVPIVPNVCIFFNMFLFGQLHHEAWVRFVILSIFMVGVYAIYGQYHA 301
>BI265362 weakly similar to GP|7296598|gb|A CG11128 gene product {alt 2}
[Drosophila melanogaster], partial (6%)
Length = 556
Score = 42.7 bits (99), Expect = 4e-04
Identities = 16/42 (38%), Positives = 25/42 (59%)
Frame = +2
Query: 564 PFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLF 605
P VP +P + +N YL++ L TW+R +WL IG ++ F
Sbjct: 230 PLVPLVPYLSVCMNVYLMVQLDYQTWVRFIIWLAIGKFLH*F 355
>TC93809 similar to PIR|H86428|H86428 hypothetical protein AAF19744.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 868
Score = 40.0 bits (92), Expect = 0.003
Identities = 38/192 (19%), Positives = 81/192 (41%), Gaps = 1/192 (0%)
Frame = +2
Query: 227 GYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCV 286
G P G + N + + F + G + ++ + +++ QR +P+G +A
Sbjct: 113 GIPEPDGSVTWNFNSLVG---LFFPAVTGIMAGSNRSSSLRDTQRSIPVGTLSATLSTSF 283
Query: 287 LYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAV-YIITTGAVTALFSSLLGSVLPQ 345
+Y L+S ++ G V D ++ + + W + +I G + + + L S+
Sbjct: 284 MY-LISVILFGAVA----TRDKLLTDRLLTATIAWPLPSLIKIGIILSTMGAALQSLTGA 448
Query: 346 PRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLL 405
PR+ A+A D +LP P +T+ T L ++ + V++ LL
Sbjct: 449 PRLLAAIANDDILPILNYFKVADGSEPHIATLFTALLCIGCVVIGNLDLITPTVTMFFLL 628
Query: 406 AFTTVAVSVLII 417
++ V +S ++
Sbjct: 629 CYSGVNLSCFLL 664
>BE942281 similar to GP|8567793|gb| amino acid transporter putative
{Arabidopsis thaliana}, partial (25%)
Length = 587
Score = 38.5 bits (88), Expect = 0.008
Identities = 18/49 (36%), Positives = 30/49 (60%)
Frame = +1
Query: 564 PFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHSS 612
P +P++PA I +N +LL L +++R V+ + VL Y+FY H+S
Sbjct: 304 PLMPWIPAISIFLNLFLLGSLDGPSYVRFGVFSAVAVLFYIFYS-VHAS 447
>BQ138029 weakly similar to GP|1764098|gb|A putative permease {Uromyces
fabae}, partial (10%)
Length = 656
Score = 37.4 bits (85), Expect = 0.018
Identities = 24/78 (30%), Positives = 38/78 (47%), Gaps = 1/78 (1%)
Frame = +2
Query: 50 IGVGATIGAGVYILIGTVAREQAGP-ALVISLFIAGIAAALSALCYAELACRCPSAGSAY 108
I +G TIG G+++ I T A AGP ++++ I G+A C E+A P G+
Sbjct: 257 IALGGTIGTGLFLGIAT-AFANAGPLSVLLGYTITGVAVFGMMQCLGEMATWLPLPGALP 433
Query: 109 HYTYICIGEGVAWLVGWS 126
Y + + VGW+
Sbjct: 434 QYCARYTDPALGFAVGWN 487
>AW774440 weakly similar to PIR|T48473|T484 amino acid transport-like protein
- Arabidopsis thaliana, partial (7%)
Length = 679
Score = 30.4 bits (67), Expect = 2.2
Identities = 14/49 (28%), Positives = 27/49 (54%)
Frame = -1
Query: 564 PFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHSS 612
PF+P+ P+ I +N +L+ L ++ R ++W + Y+ G HS+
Sbjct: 412 PFMPWPPSMSIFLNVFLMTTLKTLSFQRFAIWSCFITMFYVL*G-VHST 269
>TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (12%)
Length = 765
Score = 29.6 bits (65), Expect = 3.7
Identities = 14/41 (34%), Positives = 24/41 (58%)
Frame = +1
Query: 464 SHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALL 504
+H+ ++S + EH + K + Q+NEK +L W IAL+
Sbjct: 385 THIRNQSGIYGEH--LTKHEERFQNNEKNMERLRQWKIALI 501
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.142 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,471,251
Number of Sequences: 36976
Number of extensions: 318891
Number of successful extensions: 2203
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 2155
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2186
length of query: 636
length of database: 9,014,727
effective HSP length: 102
effective length of query: 534
effective length of database: 5,243,175
effective search space: 2799855450
effective search space used: 2799855450
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148965.10


