

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148918.3 + phase: 0
(1351 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
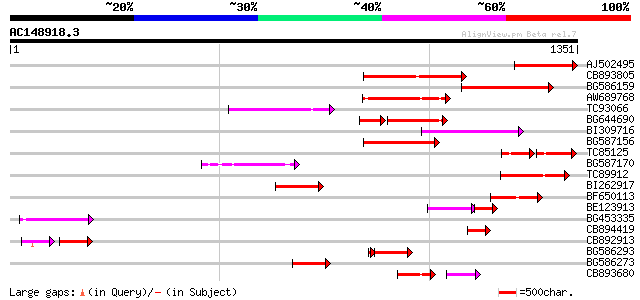
Score E
Sequences producing significant alignments: (bits) Value
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 291 1e-78
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 246 3e-65
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 241 1e-63
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 189 6e-48
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 183 4e-46
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 147 1e-45
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 179 6e-45
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 177 3e-44
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 87 6e-35
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 130 3e-30
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 129 6e-30
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 121 2e-27
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 115 9e-26
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 86 2e-24
BG453335 weakly similar to GP|18071369|g putative gag-pol polypr... 111 2e-24
CB894419 weakly similar to GP|10177935|db copia-type polyprotein... 109 6e-24
CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabi... 65 1e-22
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 100 2e-21
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 95 2e-19
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 67 5e-15
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 291 bits (745), Expect = 1e-78
Identities = 142/149 (95%), Positives = 146/149 (97%)
Frame = +2
Query: 1203 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQHVVALSTAEAEYITATSCATQTVWLRRI 1262
SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQ VVA STAEAEYI +TSCATQTVWLRRI
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1263 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIRFHKIRELIAEKEVVIEYCPT 1322
LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDI+FHKIRELIAEKEVVIEYCPT
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1323 KEQIADIFTKPLKIESFYKLKKMLGMMKA 1351
+E+IADIFTKPLKIESFYKLKKMLGMMKA
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGMMKA 448
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 246 bits (629), Expect = 3e-65
Identities = 124/249 (49%), Positives = 172/249 (68%), Gaps = 3/249 (1%)
Frame = +3
Query: 843 EPVTFEEASRDENWIKAMDEEINAIEKNKTWELTELPPDKKPIGVKWVYKTKYKPSGEID 902
+P TFEEA + E W +M+ E+ A E+N TWELT+L K IG+KW++KTK +GEI+
Sbjct: 39 DPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENGEIE 218
Query: 903 RYKARLVAKGYKQKPGIDYFEVFAPVARLDTIRMLISLSAQ---NNWKIHQMDVKSAFLN 959
+YKARLVAKGY Q+ G+DY EVFAPVAR DTIRM+I+L+AQ + I M + +
Sbjct: 219 KYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KAHSCME 398
Query: 960 GTLEEEVYVEQPAGYVVRGKEDKVYRLKKALYGLKQAPRAWYKKIDSYFIQNGFQRCPFE 1019
+ + + + + V + R+K+ALYGLKQAPRAWY +I++YF + GF++CP+E
Sbjct: 399 N*MRKFLLIN----HRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKCPYE 566
Query: 1020 HTLYIKFIDPGDVLIVCLYVDDLIFTGNNSKMIAEFRGAMISYFEMTDLGLMSYFLGIEV 1079
HTL++K + G +LI+ LYVDDLIF GN+ M EF+ +M F M+DLG M YFLG+EV
Sbjct: 567 HTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVEV 746
Query: 1080 IQQKDGIFI 1088
Q + GI+I
Sbjct: 747 TQNEKGIYI 773
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 241 bits (616), Expect = 1e-63
Identities = 113/218 (51%), Positives = 153/218 (69%)
Frame = +1
Query: 1077 IEVIQQKDGIFISQKKYASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSL 1136
+EVIQ ++GI+I Q+KY +D+L++F ME S P+ + KL ++ +G +VD+T YK +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1137 IGSLRYLTATRPDIVYGVGLLSRYMEDPCVSHLQGAKRILRYIKGTLTEGIFYGNNSDVK 1196
+G L YL ATRPD++Y + L+SR+M P H+ KR+LRY+ GT+ GI Y N K
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1197 LVGYTDSDWAGDTETRKSTSGYAFHLGTGAISWSSKKQHVVALSTAEAEYITATSCATQT 1256
L YTDSD+AGD + RKSTSGY F L +GA+SWSSKKQ VV LST +AE+I A CA Q+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1257 VWLRRILEVMHHEQNTPTKIYCDNKSAIALSKNPVFHG 1294
VW+RR+LE + + Q+ +YCDN S I LSKNPV HG
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLHG 654
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 189 bits (480), Expect = 6e-48
Identities = 94/209 (44%), Positives = 136/209 (64%)
Frame = +1
Query: 841 DCEPVTFEEASRDENWIKAMDEEINAIEKNKTWELTELPPDKKPIGVKWVYKTKYKPSGE 900
DCE + D W++AM E A+ NKTW+L LPP KK IG KWVY+ K P G
Sbjct: 64 DCENLPLS----DPRWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGS 231
Query: 901 IDRYKARLVAKGYKQKPGIDYFEVFAPVARLDTIRMLISLSAQNNWKIHQMDVKSAFLNG 960
++++KARLVAKG+ Q G DY E F+PV + TIR++++++ W+I Q+D+ +AFLNG
Sbjct: 232 VNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNG 411
Query: 961 TLEEEVYVEQPAGYVVRGKEDKVYRLKKALYGLKQAPRAWYKKIDSYFIQNGFQRCPFEH 1020
L+EEVY+ QP G+ K V +L K+LYGLKQAPRAWY+ + S IQ GF + +
Sbjct: 412 FLQEEVYMSQPQGFEAANK-SLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDP 588
Query: 1021 TLYIKFIDPGDVLIVCLYVDDLIFTGNNS 1049
+L I + G + + +YVDD++ TG+++
Sbjct: 589 SLLI-YNQNGACIYLXIYVDDILITGSSA 672
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 183 bits (464), Expect = 4e-46
Identities = 105/259 (40%), Positives = 153/259 (58%), Gaps = 7/259 (2%)
Frame = +1
Query: 522 GKKHRESFPTGKSWRAKKLLEIVHSDLCSV-EIPTPGGCRYFITFIDDFSRKAWVYFLKQ 580
G + + SF T + R K +L+ +HSDL ++ + GG RY +T IDDF RK WVYFL+
Sbjct: 43 GNRKKVSFSTA-THRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRY 219
Query: 581 KSEAVDSFKTFKAFVEKQSGCPIKALRTDRGQEYLVG--TDFFEQHGIQHQLTTRYTPQQ 638
K+E +FK ++ VE Q+G +K L TD E+ +F HGI T PQQ
Sbjct: 220 KNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQ 399
Query: 639 NGVAERKNRTIMDMVRCMLKAKQM--PKEFWAEAVATAVYILNRCPTKSVQEKTPEEAGS 696
NGVAER RT+++ RCML + ++ W EA +TA +++NR P ++ K PE+ S
Sbjct: 400 NGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWS 579
Query: 697 GRRPSIRHLRVFGCIAYAHVPDQIRKKLDDKGERCIFIGYCSNSKAYKLY--NPETKKVI 754
G +LR+FGC AYA V D KL + CIF+ Y S SK Y+L+ +P+++K+I
Sbjct: 580 GNLVDYSNLRIFGCPAYALVND---GKLAPRAGECIFLSYASESKGYRLWCSDPKSQKLI 750
Query: 755 ISRDVTFDEGGMWNWSSKS 773
+SRDVTF+E + + +S
Sbjct: 751 LSRDVTFNEDALLSSGKQS 807
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 147 bits (371), Expect(2) = 1e-45
Identities = 71/142 (50%), Positives = 106/142 (74%)
Frame = -2
Query: 901 IDRYKARLVAKGYKQKPGIDYFEVFAPVARLDTIRMLISLSAQNNWKIHQMDVKSAFLNG 960
I R K++LV +GY QK GIDY E F+PVAR++ IR+LI+ +A +K++QMDVKSAF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 961 TLEEEVYVEQPAGYVVRGKEDKVYRLKKALYGLKQAPRAWYKKIDSYFIQNGFQRCPFEH 1020
L+EEV+V+QP G+ + V+RL K LYGLKQAPRAWY+++ + ++NGF+R ++
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1021 TLYIKFIDPGDVLIVCLYVDDL 1042
TL++ ++LI+ +YVDD+
Sbjct: 64 TLFL-LKRE*ELLIIQVYVDDI 2
Score = 56.2 bits (134), Expect(2) = 1e-45
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 1/63 (1%)
Frame = -3
Query: 833 IINFALF-ADCEPVTFEEASRDENWIKAMDEEINAIEKNKTWELTELPPDKKPIGVKWVY 891
+++F+ F + EP +EA RD +WI +M EE++ E++K W L P K IG +WV+
Sbjct: 621 LVSFSAFISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVF 442
Query: 892 KTK 894
+ K
Sbjct: 441 RNK 433
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 179 bits (454), Expect = 6e-45
Identities = 96/243 (39%), Positives = 140/243 (57%)
Frame = +2
Query: 982 KVYRLKKALYGLKQAPRAWYKKIDSYFIQNGFQRCPFEHTLYIKFIDPGDVLIVCLYVDD 1041
KV L+K++YGLKQA R WY K+ I G+ + + +L+ KF D ++ +YVDD
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLL-VYVDD 193
Query: 1042 LIFTGNNSKMIAEFRGAMISYFEMTDLGLMSYFLGIEVIQQKDGIFISQKKYASDILKKF 1101
++ GN+ I + +I F++ DLG + YFLG+EV + K GI ++Q+KY ++L+
Sbjct: 194 IVLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDS 373
Query: 1102 KMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTATRPDIVYGVGLLSRYM 1161
K TP + LKL D T Y+ LIG L YLT TRPDI + V LS+++
Sbjct: 374 GNLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFV 553
Query: 1162 EDPCVSHLQGAKRILRYIKGTLTEGIFYGNNSDVKLVGYTDSDWAGDTETRKSTSGYAFH 1221
P H Q A R+L+Y+K +G+FY S++KL + DSDWA TRKS +GY
Sbjct: 554 SKPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVF 733
Query: 1222 LGT 1224
LG+
Sbjct: 734 LGS 742
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 177 bits (448), Expect = 3e-44
Identities = 85/180 (47%), Positives = 117/180 (64%)
Frame = -1
Query: 844 PVTFEEASRDENWIKAMDEEINAIEKNKTWELTELPPDKKPIGVKWVYKTKYKPSGEIDR 903
P ++EEA D+ W +++ E A+ KN TW +ELP KK + +W++ KYK G I+R
Sbjct: 558 PRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIER 379
Query: 904 YKARLVAKGYKQKPGIDYFEVFAPVARLDTIRMLISLSAQNNWKIHQMDVKSAFLNGTLE 963
K RLVA+G+ G DY E FAPVA+L TIR+++SL+ W + QMDVK+AFL G LE
Sbjct: 378 KKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELE 199
Query: 964 EEVYVEQPAGYVVRGKEDKVYRLKKALYGLKQAPRAWYKKIDSYFIQNGFQRCPFEHTLY 1023
+EVY+ P G K V RLKKA+YGLKQ+PRAWY K+ + GF++ +HTL+
Sbjct: 198 DEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 87.4 bits (215), Expect(2) = 6e-35
Identities = 45/77 (58%), Positives = 56/77 (72%)
Frame = +1
Query: 1173 KRILRYIKGTLTEGIFYGNNSDVKLVGYTDSDWAGDTETRKSTSGYAFHLGTGAISWSSK 1232
KRI+RYIKGT + +G S++ + GY DSD+AGD + RKST+GY F L GA+SW SK
Sbjct: 4 KRIMRYIKGTSGVAVCFGG-SELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSK 180
Query: 1233 KQHVVALSTAEAEYITA 1249
Q VVALST EAEY+ A
Sbjct: 181 LQTVVALSTTEAEYMAA 231
Score = 80.1 bits (196), Expect(2) = 6e-35
Identities = 32/96 (33%), Positives = 63/96 (65%)
Frame = +3
Query: 1255 QTVWLRRILEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIRFHKIRELIAEKE 1314
+ +W++R++E + H+Q T +YCD++SA+ +++NP FH R+KHI I++H +RE++ E
Sbjct: 249 EAIWMQRLMEELGHKQEQIT-VYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVEEGS 425
Query: 1315 VVIEYCPTKEQIADIFTKPLKIESFYKLKKMLGMMK 1350
V ++ T + +AD TK + + F + G+++
Sbjct: 426 VDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGLLE 533
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 130 bits (328), Expect = 3e-30
Identities = 80/235 (34%), Positives = 122/235 (51%), Gaps = 2/235 (0%)
Frame = -3
Query: 457 RLFPLRIQHDQFSCLSSIIPNDDWLWHMRFGHFHFSGLNYLSRKEYVSGLPVVKIPSGVC 516
+L P+ F+ SS+ N D LWH R GH H LN + LP V + C
Sbjct: 650 KLDPVSNYKCSFTSSSSL--NKDALWHARLGHPHGRALNLM--------LPGVVFENKNC 501
Query: 517 ETCQMGKKHRESFPTGKSWRAKKLLEIVHSDLCSVEIPTPGGCRYFITFIDDFSRKAWVY 576
E C +GK + FP S + +++++DL + + +YF+TFID+ S+ W+
Sbjct: 500 EACILGKHCKNVFPR-TSTVYENCFDLIYTDLWTAPSLSRDNHKYFVTFIDEKSKYTWLT 324
Query: 577 FLKQKSEAVDSFKTFKAFVEKQSGCPIKALRTDRGQEY--LVGTDFFEQHGIQHQLTTRY 634
+ K +D+FK F+A+V IK LR+D G EY + HGI HQ + Y
Sbjct: 323 LIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPY 144
Query: 635 TPQQNGVAERKNRTIMDMVRCMLKAKQMPKEFWAEAVATAVYILNRCPTKSVQEK 689
TPQQNGVA+RKN+ +M++ R ++ + V+TA Y++N PTK ++ K
Sbjct: 143 TPQQNGVAKRKNKHLMEVARSLM---------FQANVSTACYLINWIPTKVLRIK 6
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 129 bits (325), Expect = 6e-30
Identities = 68/165 (41%), Positives = 102/165 (61%), Gaps = 2/165 (1%)
Frame = +1
Query: 1170 QGAKRILRYIKGTLTEGIFYGNNSDVK--LVGYTDSDWAGDTETRKSTSGYAFHLGTGAI 1227
Q K +L+Y+ +L + Y + + L GY D+D+AG+ +TRKS SG+ F L I
Sbjct: 1 QALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTI 180
Query: 1228 SWSSKKQHVVALSTAEAEYITATSCATQTVWLRRILEVMHHEQNTPTKIYCDNKSAIALS 1287
SW + +Q VV LST +AEYI +WL+ ++ + Q KI+CD++SAI L+
Sbjct: 181 SWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEY-VKIHCDSQSAIHLA 357
Query: 1288 KNPVFHGRSKHIDIRFHKIRELIAEKEVVIEYCPTKEQIADIFTK 1332
+ V+H R+KHIDIR H IR++I KE+V+E ++E AD+FTK
Sbjct: 358 NHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 121 bits (303), Expect = 2e-27
Identities = 58/115 (50%), Positives = 71/115 (61%)
Frame = +1
Query: 634 YTPQQNGVAERKNRTIMDMVRCMLKAKQMPKEFWAEAVATAVYILNRCPTKSVQEKTPEE 693
+TPQQNGVAER NRT+++ R MLK M K FWAEAV TA Y++NR P+ + KTP E
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPME 261
Query: 694 AGSGRRPSIRHLRVFGCIAYAHVPDQIRKKLDDKGERCIFIGYCSNSKAYKLYNP 748
G+ L VFGC Y Q R KLD K +CIF+GY N K Y L++P
Sbjct: 262 MWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWDP 426
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 115 bits (289), Expect = 9e-26
Identities = 60/126 (47%), Positives = 81/126 (63%), Gaps = 3/126 (2%)
Frame = +1
Query: 1147 RPDIVYGVGLLSRYMEDPCVSHLQGAKRILRYIKGTLTEGIF--YGNNSDV-KLVGYTDS 1203
RPDI Y V ++S++M DP HL A RILRY++GT+ G+ YG S+V +L+ Y+DS
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 1204 DWAGDTETRKSTSGYAFHLGTGAISWSSKKQHVVALSTAEAEYITATSCATQTVWLRRIL 1263
DW GD R+STSGY F AISW +KKQ + ALS+ EAEYI T Q +WL ++
Sbjct: 301 DWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 1264 EVMHHE 1269
+ + E
Sbjct: 472 KELKCE 489
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 85.5 bits (210), Expect(2) = 2e-24
Identities = 43/118 (36%), Positives = 67/118 (56%)
Frame = +1
Query: 995 QAPRAWYKKIDSYFIQNGFQRCPFEHTLYIKFIDPGDVLIVCLYVDDLIFTGNNSKMIAE 1054
Q+PR W+ + + G+ +C +H ++IK I+ +YVDD+ TG++ K I
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1055 FRGAMISYFEMTDLGLMSYFLGIEVIQQKDGIFISQKKYASDILKKFKMEHSKPISTP 1112
+ + FE+ DLG + YFLG+EV + K G ISQ+KY D+LK+ +M K I P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 47.0 bits (110), Expect(2) = 2e-24
Identities = 23/55 (41%), Positives = 34/55 (61%)
Frame = +2
Query: 1107 KPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTATRPDIVYGVGLLSRYM 1161
KP TP++ +KL +G VD Y+ L+G L YL+ TRPDI + V +S++M
Sbjct: 338 KPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BG453335 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (11%)
Length = 660
Score = 111 bits (278), Expect = 2e-24
Identities = 58/176 (32%), Positives = 105/176 (58%)
Frame = +1
Query: 23 YWEFMMTTHLKAHNIWSYVESGLQQGADELARRRDQLALSQILQGIDYSIFGKIANAKTS 82
YW T L I ++ + + A + +++D L I +D F +I+ + S
Sbjct: 40 YWRLCKTVTL----I*VLMQPEVHRNAHKELKKKDCKELFLIQ*SLDEGNFKRISKSTRS 207
Query: 83 KEAWDILKLSHKGVEKAQKSKLQSLRREYERYEMSSSETVDQYFTRVINIVNKMRVYGED 142
KEA +IL+ H+G +K ++ KLQSLRR+++ +M + + Y +++IN+VN+++ YGE
Sbjct: 208 KEASNILEKYHEGDDKVEQIKLQSLRRKFKLMQMEDEKKIADYISKLINVVNQIKAYGEV 387
Query: 143 IQDSKVVEKILRTMPMKYDHVVTTILESHDTDTLSVAELQGSIESHVNRILEKTEK 198
+ D ++VEKI+RT+ ++D +V I ES D TL + EL+ S+E+H + E++ +
Sbjct: 388 VTDQQIVEKIMRTLSPRFDFIVVAIHESKDVKTLKIEELKSSLEAHKLMVYERSSE 555
>CB894419 weakly similar to GP|10177935|db copia-type polyprotein {Arabidopsis
thaliana}, partial (2%)
Length = 170
Score = 109 bits (273), Expect = 6e-24
Identities = 55/56 (98%), Positives = 55/56 (98%)
Frame = +3
Query: 1091 KKYASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT 1146
KK ASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT
Sbjct: 3 KKCASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT 170
>CB892913 similar to GP|1769897|emb lectin receptor kinase {Arabidopsis
thaliana}, partial (35%)
Length = 837
Score = 64.7 bits (156), Expect(2) = 1e-22
Identities = 28/78 (35%), Positives = 56/78 (70%)
Frame = +1
Query: 120 ETVDQYFTRVINIVNKMRVYGEDIQDSKVVEKILRTMPMKYDHVVTTILESHDTDTLSVA 179
E V YF+RVI+I N+++ E ++D +++EKILR++ K++H+V I E+ D +T+++
Sbjct: 472 ERVRVYFSRVISISNQLKRNSEKLEDVRIMEKILRSLDPKFEHIVEKIEETKDLETMTIE 651
Query: 180 ELQGSIESHVNRILEKTE 197
+LQGS++++ + +K +
Sbjct: 652 KLQGSLQAYEEKHKKKKD 705
Score = 61.2 bits (147), Expect(2) = 1e-22
Identities = 35/90 (38%), Positives = 51/90 (55%), Gaps = 12/90 (13%)
Frame = +3
Query: 28 MTTHLKAHNIWSYVESGLQQGADELA------------RRRDQLALSQILQGIDYSIFGK 75
M L AH++W VE G ++ DE + R+RD+ AL I Q +D F K
Sbjct: 171 MMALLGAHDVWEVVEKGYKESQDEDSLTKAQRDTLKDSRKRDKKALFLIYQALDEDEFEK 350
Query: 76 IANAKTSKEAWDILKLSHKGVEKAQKSKLQ 105
I+NA ++KEAW+ LK S +G +K +K +LQ
Sbjct: 351 ISNATSAKEAWEKLKTSCQGEDKVKKVRLQ 440
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 100 bits (249), Expect(2) = 2e-21
Identities = 48/92 (52%), Positives = 67/92 (72%)
Frame = +2
Query: 869 KNKTWELTELPPDKKPIGVKWVYKTKYKPSGEIDRYKARLVAKGYKQKPGIDYFEVFAPV 928
+ +T +L + P KPIG++W+YK K G + +YKARLVAKGY ++ GID+ EVFAPV
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
Query: 929 ARLDTIRMLISLSAQNNWKIHQMDVKSAFLNG 960
R++TI +L++L+A N IH +DVK AFLNG
Sbjct: 227 VRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
Score = 21.9 bits (45), Expect(2) = 2e-21
Identities = 9/16 (56%), Positives = 11/16 (68%)
Frame = +3
Query: 856 WIKAMDEEINAIEKNK 871
WI AM EI +I KN+
Sbjct: 9 WIDAMKAEIESIIKNR 56
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 95.1 bits (235), Expect = 2e-19
Identities = 39/90 (43%), Positives = 64/90 (70%)
Frame = -2
Query: 674 AVYILNRCPTKSVQEKTPEEAGSGRRPSIRHLRVFGCIAYAHVPDQIRKKLDDKGERCIF 733
A Y++NR PT+ ++++ P E + R+PS+ ++RVFGC+ Y VP ++R KL+ + + +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 734 IGYCSNSKAYKLYNPETKKVIISRDVTFDE 763
IGY + K YK Y+PE ++V++SRDV F E
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIE 435
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 66.6 bits (161), Expect(2) = 5e-15
Identities = 37/91 (40%), Positives = 56/91 (60%), Gaps = 1/91 (1%)
Frame = -2
Query: 925 FAPVARLDTIRMLISLSAQNNWKIHQMDVKSAFLNGTLEEEVYVEQPAGYVVR-GKEDKV 983
F P+ +L+TI L+S+ A N + +DVK+AFL G L E++Y+ QP G+ GK V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGK--MV 381
Query: 984 YRLKKALYGLKQAPRAWYKKIDSYFIQNGFQ 1014
+LKK++YGLKQ PR + + + GF+
Sbjct: 380 GKLKKSMYGLKQGPRQCI*SLKALCTRKGFR 288
Score = 33.9 bits (76), Expect(2) = 5e-15
Identities = 21/83 (25%), Positives = 42/83 (50%), Gaps = 2/83 (2%)
Frame = -3
Query: 1042 LIFTGNNSKMIAEFRGAMISYFEMTDLGLMSYFLGIEVI--QQKDGIFISQKKYASDILK 1099
L+ G+N I + +M DLG +G++++ +QK + +SQ +Y + +L+
Sbjct: 271 LLVVGSNIDEIKNLKTRFSKEIDMKDLGPAKKIIGMQIMIDKQKGVL*LSQVEYITRVLQ 92
Query: 1100 KFKMEHSKPISTPVEEKLKLTRE 1122
F M ++ +ST + L+ E
Sbjct: 91 IFNMGNAILVSTTLASHFCLSHE 23
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,885,640
Number of Sequences: 36976
Number of extensions: 641798
Number of successful extensions: 3581
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 3169
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3395
length of query: 1351
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1243
effective length of database: 5,021,319
effective search space: 6241499517
effective search space used: 6241499517
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148918.3


