

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
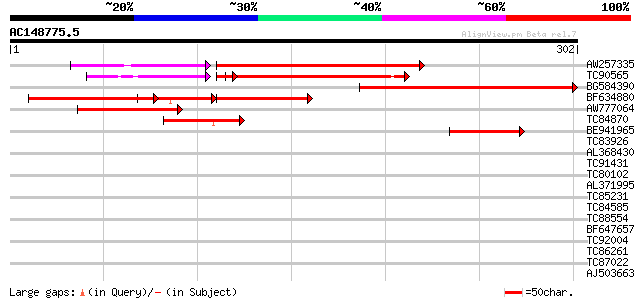
Query= AC148775.5 + phase: 0 /pseudo
(302 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW257335 weakly similar to PIR|T45719|T45 hypothetical protein F... 170 3e-51
TC90565 weakly similar to PIR|T45719|T45719 hypothetical protein... 143 1e-50
BG584390 weakly similar to GP|13161355|dbj contains ESTs AU10131... 194 3e-50
BF634880 weakly similar to GP|21554728|gb| putative esterase {Ar... 97 1e-26
AW777064 101 3e-22
TC84870 similar to GP|21554728|gb|AAM63672.1 putative esterase {... 74 7e-14
BE941965 weakly similar to GP|19699292|gb| AT4g24140/T19F6_130 {... 44 6e-05
TC83926 35 0.027
AL368430 weakly similar to GP|21592805|gb| unknown {Arabidopsis ... 34 0.079
TC91431 similar to GP|10177753|dbj|BAB11066. gene_id:MDN11.10~un... 32 0.30
TC80102 weakly similar to PIR|A86252|A86252 hypothetical protein... 32 0.30
AL371995 similar to GP|8439897|gb|A It contains Ank repeat PF|00... 32 0.39
TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {R... 30 1.5
TC84585 weakly similar to GP|15408736|dbj|BAB64139. hypothetical... 29 1.9
TC88554 weakly similar to GP|9759049|dbj|BAB09571.1 gene_id:MVA3... 29 1.9
BF647657 similar to GP|10177403|dbj gene_id:K24M7.11~unknown pro... 29 2.5
TC92004 28 5.7
TC86261 homologue to GP|22335691|dbj|BAC10548. cullin-like prote... 27 7.4
TC87022 homologue to SP|O04486|RB1C_ARATH Ras-related protein Ra... 27 9.6
AJ503663 similar to GP|8843727|dbj| UTP-glucose glucosyltransfer... 27 9.6
>AW257335 weakly similar to PIR|T45719|T45 hypothetical protein F1P2.140 -
Arabidopsis thaliana, partial (46%)
Length = 572
Score = 170 bits (431), Expect(2) = 3e-51
Identities = 75/111 (67%), Positives = 99/111 (88%)
Frame = +1
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEGSFE+GNYW+EVDD+H+VAQHF E+NR++ AI+GHSKG +VVLLYASKYH+IKTV N
Sbjct: 235 ESEGSFEFGNYWREVDDIHSVAQHFHEANRLVMAIIGHSKGANVVLLYASKYHDIKTVAN 414
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRL 221
LSGRYDLKAG+++RLGK+++++I K+GF ++K S +DY +T+ESL DRL
Sbjct: 415 LSGRYDLKAGLKDRLGKNFMKKIMKEGFIELKTKSRSVDYCITDESLKDRL 567
Score = 49.3 bits (116), Expect(2) = 3e-51
Identities = 31/75 (41%), Positives = 38/75 (50%)
Frame = +3
Query: 33 CFFSF*ALSESDNTQQKW*KTCWHIT*IFWNNNQRHCYFVSWFSLL*RYQSHIEPCCCIG 92
C F +++ N QQ W T HIT* + NQR C + S L Q H CCC
Sbjct: 18 CTKPFHPTAKNHNIQQVWK*TSRHIT*SW---NQRDCCSMPRSSSLQGRQYHDTTCCCSR 188
Query: 93 KGTD*FFPF*LFWKW 107
+ +*FFP *L WKW
Sbjct: 189 ECRN*FFPL*LHWKW 233
>TC90565 weakly similar to PIR|T45719|T45719 hypothetical protein F1P2.140 -
Arabidopsis thaliana, partial (48%)
Length = 700
Score = 143 bits (361), Expect(3) = 1e-50
Identities = 69/98 (70%), Positives = 79/98 (80%)
Frame = +1
Query: 116 FEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNLSGRY 175
F W+E +DLHAV QHF ESNR + AI+GHSKGG VVLLYASKYH+IK VVN+SGRY
Sbjct: 406 FSMATTWREAEDLHAVTQHFLESNRSVTAILGHSKGGGVVLLYASKYHDIKAVVNVSGRY 585
Query: 176 DLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVT 213
DLK G+EERLGKDY+ERI++DGF DVKR G YRVT
Sbjct: 586 DLKDGVEERLGKDYMERIKEDGFIDVKR-PGSSXYRVT 696
Score = 69.3 bits (168), Expect(3) = 1e-50
Identities = 37/66 (56%), Positives = 39/66 (59%)
Frame = +2
Query: 42 ESDNTQQKW*KTCWHIT*IFWNNNQRHCYFVSWFSLL*RYQSHIEPCCCIGKGTD*FFPF 101
ES NTQ W*K +I FWN C V W SLL R Q H E CCCIGK + FFP
Sbjct: 200 ESHNTQ*AW*KISGYIA-RFWN*GD--CNLVPWLSLLKRIQIHSEHCCCIGKSWNQFFPL 370
Query: 102 *LFWKW 107
*LFWKW
Sbjct: 371 *LFWKW 388
Score = 25.8 bits (55), Expect(3) = 1e-50
Identities = 9/11 (81%), Positives = 11/11 (99%)
Frame = +3
Query: 111 ESEGSFEYGNY 121
ES+GSF+YGNY
Sbjct: 390 ESDGSFQYGNY 422
>BG584390 weakly similar to GP|13161355|dbj contains ESTs AU101316(E10232)
C19303(E10232)~hypothetical protein~similar to
Arabidopsis thaliana, partial (30%)
Length = 633
Score = 194 bits (493), Expect = 3e-50
Identities = 92/116 (79%), Positives = 102/116 (87%)
Frame = +1
Query: 187 KDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQIDKDCRILTIHGSSD 246
+DYLERIRK+GF DVK+SSG DYRVTEESLMD L NMHE+CLQID++CR+LTIHGSSD
Sbjct: 1 QDYLERIRKEGFNDVKKSSGSFDYRVTEESLMDCLSINMHESCLQIDRECRVLTIHGSSD 180
Query: 247 EIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSFIKETIDHSKSTA 302
EI VQDAHEF+KIIPNHKLHI EGADH YNNHQDELSSV ++FI ETIDH K TA
Sbjct: 181 EINTVQDAHEFSKIIPNHKLHITEGADHLYNNHQDELSSVVINFINETIDHDKGTA 348
>BF634880 weakly similar to GP|21554728|gb| putative esterase {Arabidopsis
thaliana}, partial (23%)
Length = 681
Score = 97.4 bits (241), Expect = 6e-21
Identities = 47/70 (67%), Positives = 55/70 (78%), Gaps = 1/70 (1%)
Frame = +3
Query: 11 FLIFNINSSTK*K-FMNDLLSLICFFSF*ALSESDNTQQKW*KTCWHIT*IFWNNNQRHC 69
FL+ NI S K + FMND + FFS+*A SESDNTQQKW*KTCW+IT*+FWNN+QRHC
Sbjct: 3 FLLLNIKFSKKNREFMNDFSFIYIFFSY*ASSESDNTQQKW*KTCWNIT*MFWNNDQRHC 182
Query: 70 YFVSWFSLL* 79
Y +SW LL*
Sbjct: 183YLMSWLLLL* 212
Score = 76.6 bits (187), Expect(2) = 1e-26
Identities = 39/51 (76%), Positives = 42/51 (81%)
Frame = +2
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASK 161
ESEGSF++GN EVDDLHAVAQHFRESNRVIRAIVGHSKG + LL K
Sbjct: 404 ESEGSFKFGNQRTEVDDLHAVAQHFRESNRVIRAIVGHSKGILICLLIWKK 556
Score = 60.8 bits (146), Expect(2) = 1e-26
Identities = 30/45 (66%), Positives = 31/45 (68%), Gaps = 3/45 (6%)
Frame = +1
Query: 69 CYFVSWFSLL*RYQSH---IEPCCCIGKGTD*FFPF*LFWKWPSR 110
C F+ F LL Y H IEPCCCIGKGT *FFPF*LFWKW R
Sbjct: 283 CMFI--FLLLFSYVGH*SRIEPCCCIGKGTS*FFPF*LFWKWGKR 411
>AW777064
Length = 403
Score = 101 bits (252), Expect = 3e-22
Identities = 44/56 (78%), Positives = 50/56 (88%)
Frame = +2
Query: 37 F*ALSESDNTQQKW*KTCWHIT*IFWNNNQRHCYFVSWFSLL*RYQSHIEPCCCIG 92
+*A SESDNTQQKW*KTCW+IT*+FWNN+QRHCY +SW LL*R+ S IEPCCCIG
Sbjct: 236 Y*ASSESDNTQQKW*KTCWNIT*MFWNNDQRHCYLMSWLLLL*RH*SRIEPCCCIG 403
>TC84870 similar to GP|21554728|gb|AAM63672.1 putative esterase {Arabidopsis
thaliana}, partial (12%)
Length = 500
Score = 73.9 bits (180), Expect = 7e-14
Identities = 35/47 (74%), Positives = 37/47 (78%), Gaps = 4/47 (8%)
Frame = +1
Query: 83 SHIEPCCCIGKGTD*FFPF*LFWKW----PSRESEGSFEYGNYWKEV 125
S IEPCCCIGKGT *FFPF*LFWKW SRESEGSF++GN EV
Sbjct: 325 SRIEPCCCIGKGTS*FFPF*LFWKWLVGFSSRESEGSFKFGNQRTEV 465
>BE941965 weakly similar to GP|19699292|gb| AT4g24140/T19F6_130 {Arabidopsis
thaliana}, partial (19%)
Length = 590
Score = 44.3 bits (103), Expect = 6e-05
Identities = 15/40 (37%), Positives = 29/40 (72%)
Frame = +1
Query: 235 DCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADH 274
+C++ IHG +D++IP++ ++E K IP +L +++G DH
Sbjct: 250 NCKVTIIHGKNDDVIPLECSYEVQKRIPRAQLRVVDGKDH 369
>TC83926
Length = 555
Score = 35.4 bits (80), Expect = 0.027
Identities = 21/51 (41%), Positives = 26/51 (50%)
Frame = +1
Query: 43 SDNTQQKW*KTCWHIT*IFWNNNQRHCYFVSWFSLL*RYQSHIEPCCCIGK 93
S N QQ *+ +I ++ NQ C V L Q+H EPCCCIGK
Sbjct: 208 SHNNQQVQ*QISLYIARLW---NQEDCNMVPRLLLFKGIQNHGEPCCCIGK 351
>AL368430 weakly similar to GP|21592805|gb| unknown {Arabidopsis thaliana},
partial (25%)
Length = 491
Score = 33.9 bits (76), Expect = 0.079
Identities = 17/57 (29%), Positives = 32/57 (55%), Gaps = 2/57 (3%)
Frame = +3
Query: 238 ILTIHGSSDEIIPVQDAHEFAKIIPNH-KLHIIEGADHAYN-NHQDELSSVFMSFIK 292
+L + G D I PVQ AHE ++I +L +I+ A H +E +++ ++F++
Sbjct: 159 VLVLWGEDDNIFPVQMAHELKEVISKKARLELIKEASHVPQIEKPEEFNNIILNFLQ 329
>TC91431 similar to GP|10177753|dbj|BAB11066. gene_id:MDN11.10~unknown
protein {Arabidopsis thaliana}, partial (86%)
Length = 1047
Score = 32.0 bits (71), Expect = 0.30
Identities = 19/65 (29%), Positives = 25/65 (38%), Gaps = 2/65 (3%)
Frame = -3
Query: 60 IFWNNNQRHCYFVSWFSLL*RYQSHIEPCCC--IGKGTD*FFPF*LFWKWPSRESEGSFE 117
+FW C+F + F+ CCC + G FFP FWK
Sbjct: 559 VFWKTKPNSCHFSNHFN-----------CCCHFVSPGIHSFFP---FWK----------T 452
Query: 118 YGNYW 122
+GNYW
Sbjct: 451 FGNYW 437
>TC80102 weakly similar to PIR|A86252|A86252 hypothetical protein [imported]
- Arabidopsis thaliana, partial (69%)
Length = 1255
Score = 32.0 bits (71), Expect = 0.30
Identities = 19/101 (18%), Positives = 45/101 (43%), Gaps = 5/101 (4%)
Frame = +2
Query: 207 KLDYRVTEESLMDRLGTNMHEACLQIDKDCRILTIHGSSDEIIPVQDA-----HEFAKII 261
K +Y ++E++ + +C ++ + + + + +IP+++ H ++
Sbjct: 254 KYEYHISEDAALKAYPNCETISCSDFEEAFKAVELWLAHKVVIPIENTSGGSIHRNYDLL 433
Query: 262 PNHKLHIIEGADHAYNNHQDELSSVFMSFIKETIDHSKSTA 302
H+LHI+ A N + V F+K + HS++ A
Sbjct: 434 LRHRLHIVGEVQLATNLSLLAMPGVRKEFLKRVLSHSQALA 556
>AL371995 similar to GP|8439897|gb|A It contains Ank repeat PF|00023. EST
gb|AI996003 comes from this gene. {Arabidopsis
thaliana}, partial (16%)
Length = 424
Score = 31.6 bits (70), Expect = 0.39
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Frame = -3
Query: 137 ESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNLSGRYDL-KAGIEERLGKDYLERIRK 195
E ++ ++GG+ L A++Y + V + YDL AGI+ R G D L K
Sbjct: 221 EEGKLREIFAKQNQGGETALYVAAEYGYVDMVREMIQYYDLADAGIKARNGFDALHIAAK 42
Query: 196 DGFFDV 201
G D+
Sbjct: 41 QGDLDI 24
>TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {Rauvolfia
serpentina}, partial (34%)
Length = 1602
Score = 29.6 bits (65), Expect = 1.5
Identities = 14/43 (32%), Positives = 22/43 (50%)
Frame = +2
Query: 84 HIEPCCCIGKGTD*FFPF*LFWKWPSRESEGSFEYGNYWKEVD 126
H+ +GK T F F LFWKW + S S + ++W ++
Sbjct: 800 HVNVFKVVGKSTTKFCSFCLFWKWWNTLSR-STQRDSFWSRIE 925
>TC84585 weakly similar to GP|15408736|dbj|BAB64139. hypothetical
protein~similar to receptor serine/threonine kinase
PR5K, partial (5%)
Length = 765
Score = 29.3 bits (64), Expect = 1.9
Identities = 16/46 (34%), Positives = 23/46 (49%)
Frame = +3
Query: 168 VVNLSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVT 213
VVN++ D+K GIEE+LGK + D S G+ + T
Sbjct: 564 VVNVTVENDVKGGIEEKLGKGFRLNWIASDCSDCSXSGGRCGFXST 701
>TC88554 weakly similar to GP|9759049|dbj|BAB09571.1
gene_id:MVA3.7~pir||T13462~similar to unknown protein
{Arabidopsis thaliana}, partial (30%)
Length = 727
Score = 29.3 bits (64), Expect = 1.9
Identities = 13/39 (33%), Positives = 22/39 (56%)
Frame = +1
Query: 237 RILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHA 275
RI I G D+++P + +F P +++II ADH+
Sbjct: 388 RINVIQGDGDQVVPKECITKFKLKAPYAEINIIPNADHS 504
>BF647657 similar to GP|10177403|dbj gene_id:K24M7.11~unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 525
Score = 28.9 bits (63), Expect = 2.5
Identities = 16/31 (51%), Positives = 17/31 (54%), Gaps = 1/31 (3%)
Frame = -3
Query: 109 SRESEGSFEYGNYWKEVD-DLHAVAQHFRES 138
S ESEG FEYG K VD H + FR S
Sbjct: 106 SDESEGGFEYGRVLKSVDFAAHCIRH*FRPS 14
>TC92004
Length = 769
Score = 27.7 bits (60), Expect = 5.7
Identities = 14/45 (31%), Positives = 22/45 (48%)
Frame = +1
Query: 243 GSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVF 287
G DE +P+Q + + N KL++IEG H D + +F
Sbjct: 277 GDIDENVPIQAWMYMSNKMNNIKLNVIEGGGHYLLYRMDFMEDLF 411
>TC86261 homologue to GP|22335691|dbj|BAC10548. cullin-like protein1 {Pisum
sativum}, partial (72%)
Length = 1742
Score = 27.3 bits (59), Expect = 7.4
Identities = 19/68 (27%), Positives = 31/68 (44%), Gaps = 2/68 (2%)
Frame = +3
Query: 227 EACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAY--NNHQDELS 284
E CL+ +KD +H SS+ + + HE + N L HA ++ ++LS
Sbjct: 795 EECLKREKDRVANYLHSSSEPKLLEKVQHELLSVYANQLLEKEHSGCHALLRDDKVEDLS 974
Query: 285 SVFMSFIK 292
+F F K
Sbjct: 975 RMFRLFSK 998
>TC87022 homologue to SP|O04486|RB1C_ARATH Ras-related protein Rab11C.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (81%)
Length = 1014
Score = 26.9 bits (58), Expect = 9.6
Identities = 10/23 (43%), Positives = 13/23 (56%)
Frame = +1
Query: 68 HCYFVSWFSLL*RYQSHIEPCCC 90
HCY V+W + +S PCCC
Sbjct: 403 HCYHVNWQ----QNRSQASPCCC 459
>AJ503663 similar to GP|8843727|dbj| UTP-glucose glucosyltransferase
{Arabidopsis thaliana}, partial (17%)
Length = 550
Score = 26.9 bits (58), Expect = 9.6
Identities = 12/36 (33%), Positives = 22/36 (60%)
Frame = +1
Query: 209 DYRVTEESLMDRLGTNMHEACLQIDKDCRILTIHGS 244
+Y+V+ E + G++ HE+ ++ KDC + HGS
Sbjct: 403 EYKVSGEKALSVFGSS-HESLCKMAKDCELHLHHGS 507
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.337 0.147 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,110,869
Number of Sequences: 36976
Number of extensions: 191240
Number of successful extensions: 1251
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1245
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1249
length of query: 302
length of database: 9,014,727
effective HSP length: 96
effective length of query: 206
effective length of database: 5,465,031
effective search space: 1125796386
effective search space used: 1125796386
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148775.5


