

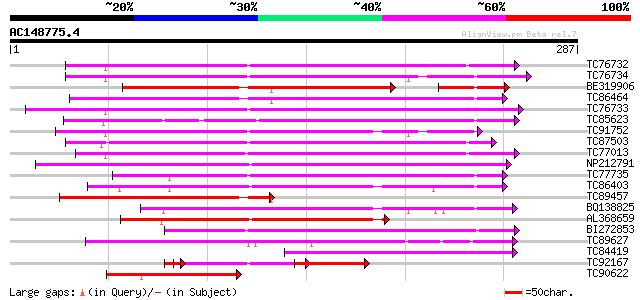
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.4 - phase: 0
(287 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 165 2e-41
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 165 2e-41
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 145 3e-40
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 160 5e-40
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 159 2e-39
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 157 3e-39
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 157 4e-39
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 144 3e-35
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 142 1e-34
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 133 9e-32
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 128 3e-30
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 123 7e-29
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 122 1e-28
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 117 5e-27
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 108 2e-24
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 96 2e-20
TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pec... 63 1e-10
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 62 2e-10
TC92167 similar to PIR|T05203|T05203 pectinesterase homolog F4I1... 47 5e-10
TC90622 weakly similar to PIR|C86366|C86366 protein F26F24.2 [im... 60 1e-09
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 165 bits (417), Expect = 2e-41
Identities = 97/233 (41%), Positives = 133/233 (56%), Gaps = 3/233 (1%)
Frame = +2
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
G FPSWV DR+LL++S + A+ VVA DGSG + + AV +AP+ K RYVI++KK
Sbjct: 683 GDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKK 862
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E++ I K+N+M++GDGM AT+ITG L++ D T + T G GF AQDI
Sbjct: 863 GTYKENIEIGKKKTNVMLVGDGMDATIITGSLNF-IDGTTTFKSATVAAVGDGFIAQDIR 1039
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
F+NTA P+ HQAVAL +D SV RC+I FQD+L A+ R + +
Sbjct: 1040FQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGN 1219
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
F K+ + RK Q+N +TAQ G E PN + Q C+V + P
Sbjct: 1220AAVVFQKSKLAARKPMANQKNMVTAQ-GREDPNQNTATSIQQCDVIPSSDLKP 1375
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 165 bits (417), Expect = 2e-41
Identities = 102/243 (41%), Positives = 139/243 (56%), Gaps = 7/243 (2%)
Frame = +3
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
G FPSWV DR+LL++S V A+ VVA DGSG + + +AV +APN RYVI++KK
Sbjct: 690 GDFPSWVTSKDRRLLESSVGDVKANVVVAKDGSGKFKTVAEAVASAPNKGTARYVIYVKK 869
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G+Y E+V I +SK+N+M++GDGM AT+ITG L++ D T T T G F AQDI
Sbjct: 870 GIYKENVEIASSKTNVMLLGDGMDATIITGSLNY-VDGTGTFQTATVAAVGDWFIAQDIG 1046
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARL-----T 201
F+NTA P+ HQAVAL SD SV RC+I FQD+L A+ R + +
Sbjct: 1047FQNTAGPQKHQAVALRVGSDRSVINRCKIDAFQDTLYAHTNRQFYRDSFITGTIDFIFGD 1226
Query: 202 SYLVRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
+ +V Q K ++ RK Q N +TAQG + PN + Q C+V + P +
Sbjct: 1227AAVVLQ----KCKLVARKPMANQNNMVTAQGRID-PNQNTATSIQQCDVIPSTDLKPVIG 1391
Query: 262 LPK 264
K
Sbjct: 1392SVK 1400
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 145 bits (367), Expect(2) = 3e-40
Identities = 80/141 (56%), Positives = 97/141 (68%), Gaps = 3/141 (2%)
Frame = +2
Query: 58 GSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGD 117
GSG+Y K+MDAV AAP S KRYVI++KKGVY E+V I K N+M+IG+GM AT+I+G
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVENVEIKKKKWNIMLIGEGMDATIISG- 178
Query: 118 LSWGRDKLDTSYTY---TFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCE 174
R+ +D S T+ TF V G GF A+DISF+NTA E HQAVAL SDSD SVFYRC
Sbjct: 179 ---SRNYVDGSTTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCG 349
Query: 175 ISGFQDSLCANIKHHSIRIAK 195
I G+QDSL + R K
Sbjct: 350 IFGYQDSLYTHTMRQFYRECK 412
Score = 37.0 bits (84), Expect(2) = 3e-40
Identities = 18/36 (50%), Positives = 23/36 (63%)
Frame = +3
Query: 218 RKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCAD 253
+KG Q+NT+TAQG + PN P GF+F F AD
Sbjct: 483 KKGMPKQKNTVTAQGRKD-PNQPTGFSFHFATFTAD 587
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 160 bits (405), Expect = 5e-40
Identities = 95/226 (42%), Positives = 128/226 (56%), Gaps = 4/226 (1%)
Frame = +1
Query: 31 FPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYN 90
+P W+ GDR+LLQ S V AD VVA+DGSGN+ + +AV AAP S KRYVI IK GVY
Sbjct: 820 WPEWISAGDRRLLQGSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYVIKIKAGVYK 999
Query: 91 EHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY---TFGVEGLGFSAQDISF 147
E+V + K+N+M +GDG T+ITG R+ +D S T+ T + G F A+DI+F
Sbjct: 1000 ENVEVPKKKTNIMFLGDGRTNTIITG----SRNVVDGSTTFHSATVAIVGGNFLARDITF 1167
Query: 148 RNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQ 207
+NTA P HQAVAL +D S FY C+I +QD+L + +
Sbjct: 1168 QNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTVDFIFGNS 1347
Query: 208 LSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCA 252
F+ DI R+ +GQ+N +TAQG + PN G Q C + A
Sbjct: 1348 AVVFQNCDIHARRPNSGQKNMVTAQGRVD-PNQNTGIVIQKCRIGA 1482
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 159 bits (401), Expect = 2e-39
Identities = 96/254 (37%), Positives = 134/254 (51%), Gaps = 2/254 (0%)
Frame = +2
Query: 9 LFCFFFRTSSSDILDTITQKGKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIM 66
+F F D T G+FPSWV DR+LL+ + + A+ VVA DGSG + +
Sbjct: 638 VFLVVFPQKDRDQFIDETLIGEFPSWVTSKDRRLLETAVGDIKANVVVAQDGSGKFKTVA 817
Query: 67 DAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLD 126
+AV +AP+ K +YVI++KKG Y E+V I + K+N+M++GDGM AT+ITG+L++ D
Sbjct: 818 EAVASAPDNGKTKYVIYVKKGTYKENVEIGSKKTNVMLVGDGMDATIITGNLNF-IDGTT 994
Query: 127 TSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANI 186
T + T G GF AQDI F+N A HQAVAL SD SV RC I FQD+L A+
Sbjct: 995 TFKSSTVAAVGDGFIAQDIWFQNMAGAAKHQAVALRVGSDQSVINRCRIDAFQDTLYAHS 1174
Query: 187 KHHSIRIAKSEARLTSYLVRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQ 246
R + + F+ LV + P QN + G E P G + Q
Sbjct: 1175NRQFYRDSVITGTIDFIFGNAAVVFQKCKLVARKPMANQNNMFTAQGREDPGQNTGTSIQ 1354
Query: 247 FCNVCADPEFLPFV 260
C++ + P V
Sbjct: 1355QCDLTPSSDLKPVV 1396
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 157 bits (398), Expect = 3e-39
Identities = 100/233 (42%), Positives = 126/233 (53%), Gaps = 2/233 (0%)
Frame = +3
Query: 28 KGKFPSWVKPGDRKLLQAS--AVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIK 85
K FP+WVKPGDRKLLQ S A A+ VVA DGSG Y + A AAP+GS RYVI++K
Sbjct: 1101 KDGFPTWVKPGDRKLLQTSSAASKANVVVAKDGSGKYTTVKAATDAAPSGSG-RYVIYVK 1277
Query: 86 KGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDI 145
GVYNE V I N+M++GDG+G T+ITG S G T + T G GF AQDI
Sbjct: 1278 AGVYNEQVEIK--AKNVMLVGDGIGKTIITGSKSVGGGTT-TFRSATVAATGDGFIAQDI 1448
Query: 146 SFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLV 205
+FRNTA NHQAVA S SD SVFY+C G+QD+L + + R +
Sbjct: 1449 TFRNTAGAANHQAVAFRSGSDLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFG 1628
Query: 206 RQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ + + P + T+TAQG + PN G V A + P
Sbjct: 1629 NAAVVLQNCNIFARNPPAKTITVTAQGRTD-PNQNTGIIIHNSRVSAQSDLNP 1784
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 157 bits (397), Expect = 4e-39
Identities = 97/228 (42%), Positives = 136/228 (59%), Gaps = 12/228 (5%)
Frame = +3
Query: 24 TITQKGKFPSWVKPGDRKLLQASA---VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRY 80
+++ + + P+W+ DRKLLQ + AD VVA DGSG Y I A+ PN S KR
Sbjct: 30 SLSHQNEEPNWLHHKDRKLLQKDSDLKKKADIVVAKDGSGKYKTISAALKHVPNKSDKRT 209
Query: 81 VIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGF 140
VI++KKG+Y E+V + +K N+M+IGDGM T+++G L++ D T T TF V G F
Sbjct: 210 VIYVKKGIYYENVRVEKTKWNVMIIGDGMNVTIVSGSLNF-IDGTPTFSTATFAVFGRNF 386
Query: 141 SAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARL 200
A+DI F+NTA P+ HQAVAL++ +D +VFY+C + +QD+L A HS R E +
Sbjct: 387 IARDIGFKNTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYA----HSNRQFYRECNI 554
Query: 201 ---------TSYLVRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNL 239
S +V Q +IL R+ GQQNTITAQG + PN+
Sbjct: 555 YGTVDFIFGNSAVVLQ----NCNILPRQPMPGQQNTITAQGKTD-PNM 683
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 144 bits (364), Expect = 3e-35
Identities = 90/222 (40%), Positives = 125/222 (55%), Gaps = 4/222 (1%)
Frame = +2
Query: 29 GKFPSWVKPGDRKLLQA---SAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIK 85
G+FP W+ +R+LL VP DAVVA DGSG Y I +A+ S +R+V+++K
Sbjct: 803 GEFPEWLGTAERRLLATVVNETVP-DAVVAKDGSGQYKTIGEALKLVKKKSLQRFVVYVK 979
Query: 86 KGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDI 145
KGVY E++ ++ + N+M+ GDGM TV++G ++ D T T TF V+G GF A+DI
Sbjct: 980 KGVYVENIDLDKNTWNVMIYGDGMTETVVSGSRNY-IDGTPTFETATFAVKGKGFIAKDI 1156
Query: 146 SFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLV 205
F NTA HQAVA+ S SD SVFYRC G+QD+L A+ R +
Sbjct: 1157 QFLNTAGASKHQAVAMRSGSDQSVFYRCSFVGYQDTLYAHSNRQFYRDCDITGTIDFIFG 1336
Query: 206 RQLSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQ 246
+ F+ I+ R+ + Q NTITAQ G + PN G Q
Sbjct: 1337 NAAAVFQNCKIMPRQPMSNQFNTITAQ-GKKDPNQNSGIVIQ 1459
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 142 bits (359), Expect = 1e-34
Identities = 88/228 (38%), Positives = 122/228 (52%), Gaps = 3/228 (1%)
Frame = +2
Query: 34 WVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNE 91
WV P DRKLLQA+ D VVA DG+GN+ I +A+ AAPN S R+VIHIK G Y E
Sbjct: 41 WVSPKDRKLLQAAVNQTKFDLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKAGAYFE 220
Query: 92 HVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTA 151
+V + K+NLM++GDG+G TV+ + D T + TF V G F A+ I+F N+A
Sbjct: 221 NVEVIKKKTNLMLVGDGIGQTVVKASRN-VVDGWTTFQSSTFAVVGDKFIAKGITFENSA 397
Query: 152 WPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSF 211
P HQAVA+ + +D S FY+C +QD+L + R + F
Sbjct: 398 GPSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNAAVVF 577
Query: 212 KT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ ++ RK Q+N TAQ G E PN G + C + A + +P
Sbjct: 578 QNCNLYARKPDPKQKNLFTAQ-GREDPNQNTGISILNCKIAAAADLIP 718
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 133 bits (334), Expect = 9e-32
Identities = 79/241 (32%), Positives = 114/241 (46%)
Frame = +1
Query: 14 FRTSSSDILDTITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAP 73
F S + + PSWV G R LL V A+AVVA DGSG + + DA+ P
Sbjct: 634 FHPSQYGVSRRLLSDDGIPSWVSDGHRHLLAGGNVKANAVVAQDGSGQFKTLTDALKTVP 813
Query: 74 NGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTF 133
+ +VI++K GVY E V + + + +IGDG T TG L++ D ++T T TF
Sbjct: 814 PTNAAPFVIYVKAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYA-DGINTYKTATF 990
Query: 134 GVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRI 193
GV G F A+DI F NTA QAVAL +D ++F+ C++ GFQD+L + R
Sbjct: 991 GVNGANFMAKDIGFENTAGTSKFQAVALRVTADQAIFHNCQMDGFQDTLFVESQRQFYRD 1170
Query: 194 AKSEARLTSYLVRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCAD 253
+ F+ L+ + P Q + GG +K N F + +
Sbjct: 1171CAISGTIDFVFGDAFGVFQNCKLICRVPAKGQKCLVTAGGRDKQNSASALVFLSSHFTGE 1350
Query: 254 P 254
P
Sbjct: 1351P 1353
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 128 bits (321), Expect = 3e-30
Identities = 77/204 (37%), Positives = 115/204 (55%), Gaps = 4/204 (1%)
Frame = +2
Query: 53 VVASDGSGNYMKIMDAVMAAPNGSKKR---YVIHIKKGVYNEHVMINNSKSNLMMIGDGM 109
+V+ G N+ I DA+ AAPN +K Y+I++++G Y E+V++ K+N++++GDG+
Sbjct: 830 LVSPYGIANHTSIGDAIAAAPNNTKPEDGYYLIYVREGYYEEYVIVPKHKNNILLVGDGI 1009
Query: 110 GATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSV 169
T+ITG+ S D T + TF V G F A DI+FRNTA PE HQAVA+ +++D S
Sbjct: 1010 NNTIITGNHSV-IDGWTTFNSSTFAVSGERFIAVDITFRNTAGPEKHQAVAVRNNADLST 1186
Query: 170 FYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSFKT-DILVRKGPTGQQNTI 228
FYRC G+QD+L + R K + F+ +I RK Q+N +
Sbjct: 1187 FYRCSFEGYQDTLYVHSVRQFYRDCKIYGTVDFIFGNAAVVFQNCNIYARKPLPNQKNAV 1366
Query: 229 TAQGGPEKPNLPFGFAFQFCNVCA 252
TAQG + PN G + Q C + A
Sbjct: 1367 TAQGRTD-PNQNTGISIQNCTIDA 1435
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 123 bits (309), Expect = 7e-29
Identities = 87/227 (38%), Positives = 121/227 (52%), Gaps = 14/227 (6%)
Frame = +3
Query: 40 RKLLQASAVPADAVV------ASDGSGNYMKIMDAVMAAPNGSKKR---YVIHIKKGVYN 90
RKLLQ++ V D VV + DGSGN+ I DA+ AAPN S ++I++ GVY
Sbjct: 714 RKLLQSNQVGEDVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSSDGYFLIYVTAGVYE 893
Query: 91 EHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNT 150
E++ I+ K+ LMMIGDG+ T+ITG+ S D T + TF V G GF +++ RNT
Sbjct: 894 EYITIDKKKTYLMMIGDGINKTIITGNHSVV-DGWTTFGSPTFAVVGQGFVGVNMTIRNT 1070
Query: 151 AWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSS 210
A HQAVAL + +D S FY C G+QD+L HS+R E + + +
Sbjct: 1071AGAVKHQAVALRNGADLSTFYSCSFEGYQDTLYT----HSLRQFYRECDIYGTVDFIFGN 1238
Query: 211 FKT-----DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCA 252
K ++ R +GQ N ITAQG + PN G + C + A
Sbjct: 1239AKVVFQNCNLYPRLPMSGQFNAITAQGRTD-PNQDTGTSIHNCTIKA 1376
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 122 bits (307), Expect = 1e-28
Identities = 64/112 (57%), Positives = 82/112 (73%), Gaps = 3/112 (2%)
Frame = +1
Query: 26 TQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIK 85
T K +FPSW++ D KLLQA+ V ADAVVA+DGSG+Y K+MDAV AAP S KRYVI++K
Sbjct: 607 TNKDRFPSWIRDEDTKLLQANGVTADAVVAADGSGDYAKVMDAVSAAPESSMKRYVIYVK 786
Query: 86 KGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY---TFG 134
KGVY E+V I K N+M+IG+GM AT+I+G R+ +D S T+ TFG
Sbjct: 787 KGVYVENVEIKKKKWNIMLIGEGMDATIISG----SRNYVDGSTTFRSATFG 930
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 117 bits (293), Expect = 5e-27
Identities = 79/202 (39%), Positives = 111/202 (54%), Gaps = 11/202 (5%)
Frame = +1
Query: 67 DAVMAAPNGS--KKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDK 124
+AV AAP+ +KR+VI+IK+GVY E V + K N++ +GDG+G TVITG + G+
Sbjct: 4 EAVNAAPDNGVDRKRFVIYIKEGVYEETVRVPLKKRNVVFLGDGIGKTVITGSANVGQPG 183
Query: 125 LDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCA 184
+ T + T V G GF A+D++ NTA P+ HQAVA DSD SV CE G QD+L A
Sbjct: 184 MTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYA 363
Query: 185 NIKHHSIRIAKSEARL---TSYLVRQLSSFKTD--ILVR----KGPTGQQNTITAQGGPE 235
HS+R R+ ++ ++ D ILVR K G+ N ITA G +
Sbjct: 364 ----HSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTD 531
Query: 236 KPNLPFGFAFQFCNVCADPEFL 257
P GF FQ C + +++
Sbjct: 532 -PAQSTGFVFQNCLINGTEDYM 594
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 108 bits (270), Expect = 2e-24
Identities = 64/139 (46%), Positives = 84/139 (60%), Gaps = 3/139 (2%)
Frame = +1
Query: 57 DGSGNYMKIMDAVMAAPNG---SKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATV 113
DGSGN+ I DA+ AAPN S ++I I +GVY E+V I+ K LMMIG+G+ T+
Sbjct: 64 DGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEYVSIDKKKKYLMMIGEGINQTI 243
Query: 114 ITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRC 173
ITG+ + D T + TF V GF A +I+FRNTA +QAVAL S +D S FY C
Sbjct: 244 ITGNRNVA-DGFTTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFYSC 420
Query: 174 EISGFQDSLCANIKHHSIR 192
G+QD+L HS+R
Sbjct: 421 SFEGYQDTLYT----HSLR 465
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 95.9 bits (237), Expect = 2e-20
Identities = 61/181 (33%), Positives = 84/181 (45%), Gaps = 1/181 (0%)
Frame = +1
Query: 79 RYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGL 138
RY I++K GVY+E++ I N++M GDG G T++TG + G + T T TF L
Sbjct: 4 RYTIYVKAGVYDEYITIPKDAVNILMYGDGPGKTIVTGRKN-GAAGVKTMQTATFANTAL 180
Query: 139 GFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEA 198
GF + ++F NTA P HQAVA + D S C I G+QD+L R
Sbjct: 181 GFIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVISG 360
Query: 199 RLTSYLVRQLSSFK-TDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFL 257
+ + + + I+VR Q NTITA G L G Q CN+
Sbjct: 361 TVDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVN-KLNTGIVIQGCNIVPKAALF 537
Query: 258 P 258
P
Sbjct: 538 P 540
>TC89627 weakly similar to GP|17529290|gb|AAL38872.1 putative pectin
methylesterase {Arabidopsis thaliana}, partial (33%)
Length = 1289
Score = 62.8 bits (151), Expect = 1e-10
Identities = 62/229 (27%), Positives = 100/229 (43%), Gaps = 10/229 (4%)
Frame = +3
Query: 39 DRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNS 98
D KL +A + V+ DGS + I +A+ + + +R +I I G Y E +++ +
Sbjct: 198 DPKLKKAESNKVRLKVSQDGSAQFKSITEALNSIQPYNIRRVIISIAPGYYREKIVVPKT 377
Query: 99 KSNLMMIGDGMGATVITGDLS---WGRD--KLDTSYTYTFGVEGLGFSAQDISFRNTA-- 151
+ +GD ITG+ + G D +L T + T V F A +I+F NTA
Sbjct: 378 LPFITFLGDVRDPPTITGNDTQSVTGSDGAQLRTFNSATVAVNASYFMAININFENTASF 557
Query: 152 --WPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLS 209
+ QAVA+ + + FY C SG QD+L + H + + ++
Sbjct: 558 PIGSKVEQAVAVRITGNKTAFYNCTFSGVQDTLYDHKGLHYFNNCTIKGSV-DFICGHGK 734
Query: 210 SFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCAD-PEFL 257
S +R +ITAQ G P+ GF+F+ V D P +L
Sbjct: 735 SLYEGCTIR-SIANNMTSITAQSG-SNPSYDSGFSFKNSMVIGDGPTYL 875
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 62.4 bits (150), Expect = 2e-10
Identities = 34/118 (28%), Positives = 55/118 (45%)
Frame = +2
Query: 140 FSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEAR 199
F+A ++ F N+A HQAVAL +D ++FY C+++G+QD+L K R
Sbjct: 86 FTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCTITGT 265
Query: 200 LTSYLVRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFL 257
+ + F+ L+ + P Q + GG K + FQ C+ +PE L
Sbjct: 266 IDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGEPEVL 439
>TC92167 similar to PIR|T05203|T05203 pectinesterase homolog F4I10.160 -
Arabidopsis thaliana, partial (11%)
Length = 652
Score = 47.4 bits (111), Expect(3) = 5e-10
Identities = 21/38 (55%), Positives = 25/38 (65%)
Frame = +1
Query: 145 ISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSL 182
+ FRNTA PE HQAVA +D +VF C GFQD+L
Sbjct: 214 VGFRNTAGPEGHQAVAARVQADRAVFANCRFEGFQDTL 327
Score = 29.6 bits (65), Expect(3) = 5e-10
Identities = 19/69 (27%), Positives = 33/69 (47%)
Frame = +3
Query: 84 IKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQ 143
+KK +E V I + ++ + GDG ++ITG ++ RD + T + V G GF
Sbjct: 33 LKKECTDEVVTITDKMKDITIYGDGSQKSIITGSKNF-RDGVTNINTASLVVLGEGFLGL 209
Query: 144 DISFRNTAW 152
+ +W
Sbjct: 210 ASGIQKHSW 236
Score = 23.1 bits (48), Expect(3) = 5e-10
Identities = 8/11 (72%), Positives = 11/11 (99%)
Frame = +2
Query: 79 RYVIHIKKGVY 89
RYVI++K+GVY
Sbjct: 17 RYVIYVKEGVY 49
>TC90622 weakly similar to PIR|C86366|C86366 protein F26F24.2 [imported] -
Arabidopsis thaliana, partial (10%)
Length = 537
Score = 59.7 bits (143), Expect = 1e-09
Identities = 30/71 (42%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Frame = +1
Query: 50 ADAVVASDGSGNYMKI---MDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIG 106
AD VA DGSG++ I +DA+ + + R VIH+K GVY+E V I N+M +G
Sbjct: 271 ADFTVAQDGSGSHRTIKEAVDALASMGHNRPSRAVIHVKAGVYHEKVEIEKKLHNVMFVG 450
Query: 107 DGMGATVITGD 117
DG+ T++TG+
Sbjct: 451 DGIDKTIVTGN 483
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,020,506
Number of Sequences: 36976
Number of extensions: 120898
Number of successful extensions: 797
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 765
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 768
length of query: 287
length of database: 9,014,727
effective HSP length: 95
effective length of query: 192
effective length of database: 5,502,007
effective search space: 1056385344
effective search space used: 1056385344
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148775.4


