

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.1 - phase: 0
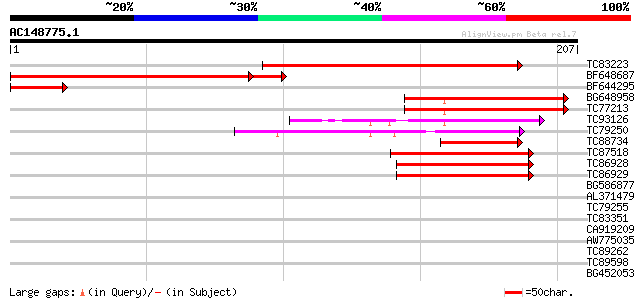
(207 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83223 similar to GP|8777436|dbj|BAA97026.1 gb|AAC78547.1~gene_... 195 1e-50
BF648687 similar to GP|2289857|gb|A Similiar to mucin and severa... 175 2e-48
BF644295 45 2e-05
BG648958 similar to GP|19310467|gb| At1g01260/F6F3_25 {Arabidops... 45 2e-05
TC77213 similar to PIR|T10861|T10861 phaseolin G-box binding pro... 44 3e-05
TC93126 44 3e-05
TC79250 similar to PIR|T06329|T06329 symbiotic ammonium transpor... 42 2e-04
TC88734 similar to GP|13346180|gb|AAK19612.1 GHDEL61 {Gossypium ... 40 6e-04
TC87518 weakly similar to PIR|E96714|E96714 probable DNA-binding... 40 6e-04
TC86928 similar to PIR|E96714|E96714 probable DNA-binding protei... 40 8e-04
TC86929 similar to PIR|T50498|T50498 myc-like protein - Arabidop... 40 8e-04
BG586877 similar to GP|16226919|gb AT4g16430/dl4240w {Arabidopsi... 39 0.002
AL371479 similar to PIR|E96714|E967 probable DNA-binding protein... 38 0.003
TC79255 similar to PIR|E96714|E96714 probable DNA-binding protei... 38 0.003
TC83351 similar to PIR|T47788|T47788 hypothetical protein F17J16... 37 0.004
CA919209 similar to SP|O80536|PIF3_ Phytochrome-interacting fact... 36 0.009
AW775035 similar to GP|13486760|dbj hypothetical protein {Oryza ... 36 0.009
TC89262 similar to GP|21537346|gb|AAM61687.1 unknown {Arabidopsi... 36 0.012
TC89598 weakly similar to GP|20127014|gb|AAM10934.1 putative bHL... 35 0.027
BG452053 homologue to GP|11994233|db contains similarity to heli... 35 0.027
>TC83223 similar to GP|8777436|dbj|BAA97026.1
gb|AAC78547.1~gene_id:MHM17.7~similar to unknown protein
{Arabidopsis thaliana}, partial (13%)
Length = 514
Score = 195 bits (495), Expect = 1e-50
Identities = 95/95 (100%), Positives = 95/95 (100%)
Frame = +1
Query: 93 HQPQQNYVAHPEASAFGRYRHDKSPNIGSNFRRQSLMKRSFAFFRSLNLMRMRDRNQAAR 152
HQPQQNYVAHPEASAFGRYRHDKSPNIGSNFRRQSLMKRSFAFFRSLNLMRMRDRNQAAR
Sbjct: 1 HQPQQNYVAHPEASAFGRYRHDKSPNIGSNFRRQSLMKRSFAFFRSLNLMRMRDRNQAAR 180
Query: 153 PSSNQLHHMISERRRREKLNENFQALRALLPQGTK 187
PSSNQLHHMISERRRREKLNENFQALRALLPQGTK
Sbjct: 181 PSSNQLHHMISERRRREKLNENFQALRALLPQGTK 285
>BF648687 similar to GP|2289857|gb|A Similiar to mucin and several other
Ser-Thr-rich proteins {Saccharomyces cerevisiae},
partial (3%)
Length = 591
Score = 175 bits (444), Expect(2) = 2e-48
Identities = 87/89 (97%), Positives = 87/89 (97%)
Frame = +1
Query: 1 MRSLSTAGSPEYSSLIFNIPPGTSPPQFSPDLTQLGGVNIPPMRPVSNTLLPLQLQQLPQ 60
MRSLSTAGSPEYSSLIFNIPPGTS PQFSPDLTQLGGVNIPPMRPVSNTLLPLQLQQLPQ
Sbjct: 286 MRSLSTAGSPEYSSLIFNIPPGTSXPQFSPDLTQLGGVNIPPMRPVSNTLLPLQLQQLPQ 465
Query: 61 ITPTQLYPTTQIENDVIMRAIQNVLSTPP 89
ITPTQLYPTTQIENDVIMRAI NVLSTPP
Sbjct: 466 ITPTQLYPTTQIENDVIMRAIPNVLSTPP 552
Score = 33.9 bits (76), Expect(2) = 2e-48
Identities = 13/14 (92%), Positives = 13/14 (92%)
Frame = +2
Query: 88 PPSHQHQPQQNYVA 101
PP HQHQPQQNYVA
Sbjct: 548 PPFHQHQPQQNYVA 589
>BF644295
Length = 458
Score = 45.4 bits (106), Expect = 2e-05
Identities = 21/21 (100%), Positives = 21/21 (100%)
Frame = +3
Query: 1 MRSLSTAGSPEYSSLIFNIPP 21
MRSLSTAGSPEYSSLIFNIPP
Sbjct: 396 MRSLSTAGSPEYSSLIFNIPP 458
>BG648958 similar to GP|19310467|gb| At1g01260/F6F3_25 {Arabidopsis
thaliana}, partial (15%)
Length = 806
Score = 45.1 bits (105), Expect = 2e-05
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Frame = +3
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + +P++ + L+H+ +ER+RREKLN+ F ALRA++P +K+ L+ I
Sbjct: 279 RPRKRGRKPANGRDEPLNHVEAERQRREKLNQRFYALRAVVPNISKMDKASLLGDAIAYI 458
Query: 202 NAL 204
N L
Sbjct: 459 NEL 467
>TC77213 similar to PIR|T10861|T10861 phaseolin G-box binding protein PG1 -
kidney bean, partial (70%)
Length = 2391
Score = 44.3 bits (103), Expect = 3e-05
Identities = 23/63 (36%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Frame = +3
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R + + +P++ + L+H+ +ER+RREKLN+ F ALRA++P +K+ L+ I
Sbjct: 1494 RPKKRGRKPANGREEPLNHVEAERQRREKLNQKFYALRAVVPNVSKMDKASLLGDAISYI 1673
Query: 202 NAL 204
N L
Sbjct: 1674 NEL 1682
>TC93126
Length = 670
Score = 44.3 bits (103), Expect = 3e-05
Identities = 41/104 (39%), Positives = 51/104 (48%), Gaps = 11/104 (10%)
Frame = +1
Query: 103 PEASAFGRYRHDKSPNIGSNFRRQSLMK---------RSFAFFR-SLNLMRMRDRNQAAR 152
P SAF +YR PNI +F QS +K R AF R SL L +RN+ A
Sbjct: 121 PNLSAFVQYRDQ--PNI--SFGEQSFLKGSNSNNMNKRMIAFLRKSLPL----ERNKVAE 276
Query: 153 PSSNQ-LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLIS 195
+ HMISER RR++ + L A+LP GTKV K S
Sbjct: 277 CERERGFKHMISERMRRQRQRQCCFNLHAVLPHGTKVYIKTFTS 408
>TC79250 similar to PIR|T06329|T06329 symbiotic ammonium transport protein
SAT1 - soybean, partial (34%)
Length = 1145
Score = 42.0 bits (97), Expect = 2e-04
Identities = 37/120 (30%), Positives = 56/120 (45%), Gaps = 14/120 (11%)
Frame = +2
Query: 83 NVLSTPPSHQHQPQ--QNYVAHPEASAFGRYRHDKSPNIGSNFRRQSLMK---RSFAFFR 137
+V T PS+ P + +H FG + NIGS + + + +S A
Sbjct: 248 DVNKTNPSNIGYPSLPKKDSSHSYILCFGNENPESMLNIGSTLKPKGKVSNHGKSLASKG 427
Query: 138 SL---------NLMRMRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKV 188
SL N+ + + AAR N H+I+ER+RREK+++ F AL ALLP K+
Sbjct: 428 SLENQKKGPKRNIQESKKTDSAAR---NAQDHIIAERKRREKISQKFIALSALLPDLKKM 598
>TC88734 similar to GP|13346180|gb|AAK19612.1 GHDEL61 {Gossypium hirsutum},
partial (40%)
Length = 1701
Score = 40.0 bits (92), Expect = 6e-04
Identities = 16/30 (53%), Positives = 24/30 (79%)
Frame = +3
Query: 158 LHHMISERRRREKLNENFQALRALLPQGTK 187
++H++SERRRR KLNE F LR+++P +K
Sbjct: 1401 MNHVLSERRRRAKLNERFLTLRSMVPSNSK 1490
>TC87518 weakly similar to PIR|E96714|E96714 probable DNA-binding protein
T6L1.19 [imported] - Arabidopsis thaliana, partial (41%)
Length = 1337
Score = 40.0 bits (92), Expect = 6e-04
Identities = 19/52 (36%), Positives = 32/52 (61%)
Frame = +2
Query: 140 NLMRMRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKVRTK 191
N +++ + + R + ++ H +E+RRR K+NE FQALR L+P+ R K
Sbjct: 149 NNVKVDEPIRGKRANPHRSKHSETEQRRRSKINERFQALRDLIPENDSKRDK 304
>TC86928 similar to PIR|E96714|E96714 probable DNA-binding protein T6L1.19
[imported] - Arabidopsis thaliana, partial (39%)
Length = 1926
Score = 39.7 bits (91), Expect = 8e-04
Identities = 17/50 (34%), Positives = 31/50 (62%)
Frame = +3
Query: 142 MRMRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKVRTK 191
+R+ ++ +P++ + H +E+RRR K+N+ FQ LR L+P + R K
Sbjct: 525 VRVDGKSTDQKPNTPRSKHSATEQRRRSKINDRFQMLRELIPHSDQKRDK 674
>TC86929 similar to PIR|T50498|T50498 myc-like protein - Arabidopsis thaliana,
partial (14%)
Length = 1289
Score = 39.7 bits (91), Expect = 8e-04
Identities = 17/50 (34%), Positives = 31/50 (62%)
Frame = +3
Query: 142 MRMRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKVRTK 191
+R+ ++ +P++ + H +E+RRR K+N+ FQ LR L+P + R K
Sbjct: 1005 VRVDGKSTDQKPNTPRSKHSATEQRRRSKINDRFQMLRELIPHSDQKRDK 1154
>BG586877 similar to GP|16226919|gb AT4g16430/dl4240w {Arabidopsis thaliana},
partial (35%)
Length = 755
Score = 38.5 bits (88), Expect = 0.002
Identities = 19/47 (40%), Positives = 33/47 (69%), Gaps = 3/47 (6%)
Frame = +1
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKV 188
+ R + +P++ + L+H+ +ER+RREKLN+ F ALRA +P +K+
Sbjct: 544 KPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAGVPNISKM 684
>AL371479 similar to PIR|E96714|E967 probable DNA-binding protein T6L1.19
[imported] - Arabidopsis thaliana, partial (26%)
Length = 423
Score = 37.7 bits (86), Expect = 0.003
Identities = 17/40 (42%), Positives = 25/40 (62%)
Frame = +1
Query: 145 RDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQ 184
+D + S + H ++E+RRR K+NE FQ LR L+PQ
Sbjct: 184 KDGKATDKASVIRSKHSVTEQRRRSKINERFQILRDLIPQ 303
>TC79255 similar to PIR|E96714|E96714 probable DNA-binding protein T6L1.19
[imported] - Arabidopsis thaliana, partial (30%)
Length = 847
Score = 37.7 bits (86), Expect = 0.003
Identities = 17/40 (42%), Positives = 25/40 (62%)
Frame = +2
Query: 145 RDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQ 184
+D + S + H ++E+RRR K+NE FQ LR L+PQ
Sbjct: 260 KDGKATDKASVIRSKHSVTEQRRRSKINERFQILRDLIPQ 379
>TC83351 similar to PIR|T47788|T47788 hypothetical protein F17J16.110 -
Arabidopsis thaliana, partial (18%)
Length = 882
Score = 37.4 bits (85), Expect = 0.004
Identities = 17/41 (41%), Positives = 28/41 (67%)
Frame = +1
Query: 147 RNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTK 187
R+ +AR + H +SERRRR+++NE +AL+ L+P +K
Sbjct: 49 RSGSARRNRAAEVHNLSERRRRDRINEKMKALQQLIPHSSK 171
>CA919209 similar to SP|O80536|PIF3_ Phytochrome-interacting factor 3
(Phytochrome-associated protein 3), partial (11%)
Length = 727
Score = 36.2 bits (82), Expect = 0.009
Identities = 15/29 (51%), Positives = 22/29 (75%)
Frame = -2
Query: 160 HMISERRRREKLNENFQALRALLPQGTKV 188
H +SERRRR+++NE +AL+ L+P KV
Sbjct: 342 HNLSERRRRDRINEKMRALQELIPNCNKV 256
>AW775035 similar to GP|13486760|dbj hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 716
Score = 36.2 bits (82), Expect = 0.009
Identities = 15/29 (51%), Positives = 22/29 (75%)
Frame = +1
Query: 160 HMISERRRREKLNENFQALRALLPQGTKV 188
H +SERRRR+++NE +AL+ L+P KV
Sbjct: 508 HNLSERRRRDRINEKMRALQELIPNCNKV 594
>TC89262 similar to GP|21537346|gb|AAM61687.1 unknown {Arabidopsis
thaliana}, partial (40%)
Length = 1198
Score = 35.8 bits (81), Expect = 0.012
Identities = 17/51 (33%), Positives = 34/51 (66%), Gaps = 1/51 (1%)
Frame = +3
Query: 144 MRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKV-RTKLL 193
M +N++ + ++++ERRRR++LN+ LRA++P+ +K+ RT +L
Sbjct: 510 MERKNRSKKLQGQPSKNLMAERRRRKRLNDRLSMLRAIVPKISKMDRTAIL 662
>TC89598 weakly similar to GP|20127014|gb|AAM10934.1 putative bHLH
transcription factor {Arabidopsis thaliana}, partial
(24%)
Length = 849
Score = 34.7 bits (78), Expect = 0.027
Identities = 18/35 (51%), Positives = 25/35 (71%)
Frame = +3
Query: 160 HMISERRRREKLNENFQALRALLPQGTKVRTKLLI 194
H+I+ER+RREKL++ F AL ++LP G K K I
Sbjct: 540 HVIAERKRREKLSQRFIALSSILP-GLKKMDKATI 641
>BG452053 homologue to GP|11994233|db contains similarity to helix-loop-helix
DNA-binding protein~gene_id:MUJ8.4 {Arabidopsis
thaliana}, partial (27%)
Length = 613
Score = 34.7 bits (78), Expect = 0.027
Identities = 18/59 (30%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Frame = +1
Query: 127 SLMKRSFAFFRSLNLMRMRDRN--QAARPSSNQLHHMISERRRREKLNENFQALRALLP 183
S+ +++ A + N R R R + S ++ H+ ER RR+++NE+ + LR+L+P
Sbjct: 199 SVQEKNCAVVQENNKKRKRPRTVKTSEEVESQRMTHIAVERNRRKQMNEHLRVLRSLMP 375
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,636,861
Number of Sequences: 36976
Number of extensions: 93284
Number of successful extensions: 814
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 800
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 812
length of query: 207
length of database: 9,014,727
effective HSP length: 92
effective length of query: 115
effective length of database: 5,612,935
effective search space: 645487525
effective search space used: 645487525
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148775.1


