

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148762.7 + phase: 2 /partial
(212 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
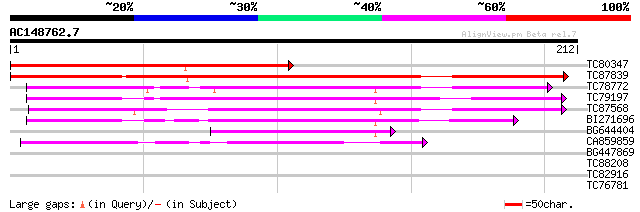
Score E
Sequences producing significant alignments: (bits) Value
TC80347 similar to GP|6714467|gb|AAF26153.1| unknown protein {Ar... 209 8e-55
TC87839 similar to GP|20259677|gb|AAM14356.1 unknown protein {Ar... 196 7e-51
TC78772 similar to GP|21593919|gb|AAM65884.1 unknown {Arabidopsi... 107 3e-24
TC79197 similar to PIR|C71447|C71447 hypothetical protein - Arab... 105 1e-23
TC87568 similar to GP|21593540|gb|AAM65507.1 putative splicing r... 105 1e-23
BI271696 weakly similar to GP|21593120|gb putative splicing regu... 92 2e-19
BG644404 similar to GP|21593919|gb unknown {Arabidopsis thaliana... 52 1e-07
CA859859 40 9e-04
BG447869 weakly similar to GP|18071362|gb| putative splicing reg... 30 0.68
TC88208 similar to GP|10177672|dbj|BAB11032. glucosidase II alph... 27 5.8
TC82916 weakly similar to GP|20067231|gb|AAM09563.1 REP13 {Secal... 27 7.5
TC76781 similar to GP|12539609|gb|AAG59584.1 pathogen-inducible ... 26 9.8
>TC80347 similar to GP|6714467|gb|AAF26153.1| unknown protein {Arabidopsis
thaliana}, partial (26%)
Length = 606
Score = 209 bits (531), Expect = 8e-55
Identities = 106/109 (97%), Positives = 106/109 (97%), Gaps = 3/109 (2%)
Frame = +1
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSI 60
SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSI
Sbjct: 280 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSI 459
Query: 61 TYSQ---MDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDE 106
TYSQ MDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDE
Sbjct: 460 TYSQDMHMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDE 606
>TC87839 similar to GP|20259677|gb|AAM14356.1 unknown protein {Arabidopsis
thaliana}, partial (84%)
Length = 1160
Score = 196 bits (497), Expect = 7e-51
Identities = 105/215 (48%), Positives = 149/215 (68%), Gaps = 6/215 (2%)
Frame = +1
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSI 60
S DY TAYVTF+DAY+ +TA LL+G+ ILD+ + I+RW Y ++ + W+ + +HE+
Sbjct: 265 SGDYACTAYVTFKDAYSQDTACLLSGATILDQRVCITRWGQY-EEHDFWSRPSYSHEEEN 441
Query: 61 TYSQM------DKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTA 114
+Y+ +FVSS GEA+ M Q+VVKTM+AKG+VLSKDA AK FDES VS+TA
Sbjct: 442 SYTTQHQTQHSSQFVSSAGEAVAMTQEVVKTMLAKGYVLSKDALSKAKDFDESHRVSATA 621
Query: 115 ANKVAELSNKIGLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAM 174
KV+ELS KIGLT+ +++G + KSVD+KY+V+ T +A A+ TGR
Sbjct: 622 TAKVSELSQKIGLTDKLSAGYDAVKSVDQKYNVSGTTMAA-----------ASATGRTVA 768
Query: 175 AAGSAIANSSYFAKGALWVSDMLSRAAKSTADLGH 209
AA +++ NSSYF+KGALW+S L+RAA++ +DLG+
Sbjct: 769 AAANSVVNSSYFSKGALWMSGALTRAAQAASDLGN 873
>TC78772 similar to GP|21593919|gb|AAM65884.1 unknown {Arabidopsis
thaliana}, partial (87%)
Length = 1409
Score = 107 bits (268), Expect = 3e-24
Identities = 74/211 (35%), Positives = 111/211 (52%), Gaps = 14/211 (6%)
Frame = +3
Query: 7 TAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWN--------NLTPNHED 58
TA+VTF+D ALE ALLL+G+ I+D+ +SIS E Y + N+ P ED
Sbjct: 231 TAFVTFKDPRALEIALLLSGATIVDQIVSISPVENYVANRETQEVGVVEYAINIAP--ED 404
Query: 59 SITYSQMDKFVSSPGEA--LTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAAN 116
+ + ++ +K PG L+ AQ VV TM+AKG + +DA AKAFDE +++ A+
Sbjct: 405 AASNTEEEK----PGGRIYLSKAQDVVTTMLAKGSAIRQDAVNKAKAFDEKHQLTANASA 572
Query: 117 KVAELSNKIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRA 172
KV+ ++GLTE + G+ E KSVD++ HV+D T +A R
Sbjct: 573 KVSNFDKRVGLTEKLTVGLSVVNEKVKSVDQRLHVSDKTMAA-----------IFAAERK 719
Query: 173 AMAAGSAIANSSYFAKGALWVSDMLSRAAKS 203
GSA+ S Y G W++ S+ A++
Sbjct: 720 LNDTGSAVKTSRYVTAGTTWLTGAFSKVARA 812
>TC79197 similar to PIR|C71447|C71447 hypothetical protein - Arabidopsis
thaliana, partial (74%)
Length = 1015
Score = 105 bits (262), Expect = 1e-23
Identities = 71/206 (34%), Positives = 105/206 (50%), Gaps = 4/206 (1%)
Frame = +2
Query: 7 TAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMD 66
TAYVTF+D+ ETA+LL+G+ I+D ++I+ Y L P E ++
Sbjct: 329 TAYVTFKDSQGAETAVLLSGATIVDLSVNITLDPDY--------QLPP--EALVSPVSES 478
Query: 67 KFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIG 126
K AL A+ VV +M AKGF+L KDA AK FDE +SS A+ VA + K+G
Sbjct: 479 KTPGGADSALRKAEDVVTSMAAKGFILGKDAVNKAKTFDEKLQLSSKASATVASVDQKLG 658
Query: 127 LTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIAN 182
L++ I +G + + VD+K+ V++ TKSA T AGSAI
Sbjct: 659 LSDKIGAGASVVSDKVREVDQKFLVSEKTKSAFAAAEQT-----------VSTAGSAIMK 805
Query: 183 SSYFAKGALWVSDMLSRAAKSTADLG 208
+ Y GA WV+ ++ +K ++G
Sbjct: 806 NRYVLTGASWVTGAFNKVSKGAVEVG 883
>TC87568 similar to GP|21593540|gb|AAM65507.1 putative splicing regulatory
protein {Arabidopsis thaliana}, partial (75%)
Length = 1188
Score = 105 bits (262), Expect = 1e-23
Identities = 72/210 (34%), Positives = 102/210 (48%), Gaps = 9/210 (4%)
Frame = +2
Query: 8 AYVTFRDAYALETALLLNGSMILDRCISISRWETYTDD-----SNNWNNLTPNHEDSITY 62
A+VTF+D ETA+LL+G+ I+D + I+ Y S+ TP DS
Sbjct: 260 AFVTFKDPQGAETAVLLSGATIVDLSVKITLDPDYKLPPAALASSASEGKTPGGADS--- 430
Query: 63 SQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELS 122
AL A+ VV +M+AKGF+L KDA AK FDE +SSTA+ KV
Sbjct: 431 ------------ALRKAEDVVTSMLAKGFILGKDAVNKAKGFDEKHQLSSTASAKVTSFD 574
Query: 123 NKIGLTETINSGIET----FKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGS 178
K+GL+E + +G K VD+K+ V++ TKSA + AGS
Sbjct: 575 QKLGLSEKLTAGASVVSGRVKEVDQKFQVSEKTKSA-----------FAAAEQKVSTAGS 721
Query: 179 AIANSSYFAKGALWVSDMLSRAAKSTADLG 208
AI + Y G WV+ ++ AK+ D+G
Sbjct: 722 AIMKNRYILTGTTWVTGAFNKVAKAAGDVG 811
>BI271696 weakly similar to GP|21593120|gb putative splicing regulatory
protein {Arabidopsis thaliana}, partial (66%)
Length = 690
Score = 91.7 bits (226), Expect = 2e-19
Identities = 67/188 (35%), Positives = 99/188 (52%), Gaps = 4/188 (2%)
Frame = +2
Query: 7 TAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMD 66
TAYVTF+++ +TA+LL GS I + ++I+ +E Y L P + +S
Sbjct: 104 TAYVTFKNSQGADTAVLLTGSNIANSPVTITPFENY--------QLPPEAQP---FSPNQ 250
Query: 67 KFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIG 126
K + A+ A+ V TM+AKGFVL K A AK+FDE + S A+ VA + KIG
Sbjct: 251 KQTPA---AVKKAEDAVSTMLAKGFVLGKGAINRAKSFDERHHLISNASATVASIDRKIG 421
Query: 127 LTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIAN 182
LT+ ++ G E + +DEK+ V++ TKS V V + A AGSAI +
Sbjct: 422 LTDKLSIGTAIVNEKVREMDEKFQVSEKTKS-----------VYAVAEQKASDAGSAIMS 568
Query: 183 SSYFAKGA 190
+ Y + GA
Sbjct: 569 NPYVSTGA 592
>BG644404 similar to GP|21593919|gb unknown {Arabidopsis thaliana}, partial
(46%)
Length = 478
Score = 52.4 bits (124), Expect = 1e-07
Identities = 28/73 (38%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Frame = +1
Query: 76 LTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIGLTETINSGI 135
++ AQ VV ++AKG + +DA AKAFDE + +TA+ +V ++GLTE + GI
Sbjct: 172 ISKAQDVVSNVLAKGSAIRQDAMNKAKAFDEKHQLRATASARVISFDRRVGLTEKLTVGI 351
Query: 136 ----ETFKSVDEK 144
E KS D++
Sbjct: 352 SVVNEEVKSGDQR 390
>CA859859
Length = 628
Score = 39.7 bits (91), Expect = 9e-04
Identities = 39/152 (25%), Positives = 64/152 (41%)
Frame = +1
Query: 5 ESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQ 64
+ TA+VTF A +TAL+L+ S++ I++ E Y S + E Q
Sbjct: 13 KQTAHVTFEKAEGAKTALMLSNSVLGSNPITVKLDENYLSTSEE------SSEPGDDIEQ 174
Query: 65 MDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNK 124
DK P A ++ ++A G+ L+ FD S +S+ + +L N
Sbjct: 175 EDK----PKAA------ILAEILAAGYQLTDSIIQRGIDFDASIGLSARVKEYLNKLQNN 324
Query: 125 IGLTETINSGIETFKSVDEKYHVTDITKSAAT 156
I KS+DE+Y V++ + AT
Sbjct: 325 I-------------KSLDEQYKVSETVTAKAT 381
>BG447869 weakly similar to GP|18071362|gb| putative splicing regulatory
protein {Oryza sativa}, partial (20%)
Length = 629
Score = 30.0 bits (66), Expect = 0.68
Identities = 12/21 (57%), Positives = 17/21 (80%)
Frame = -2
Query: 7 TAYVTFRDAYALETALLLNGS 27
TAYVTF+D + E A+LL+G+
Sbjct: 463 TAYVTFKDTHGAEXAVLLSGT 401
>TC88208 similar to GP|10177672|dbj|BAB11032. glucosidase II alpha subunit
{Arabidopsis thaliana}, partial (34%)
Length = 1505
Score = 26.9 bits (58), Expect = 5.8
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = +1
Query: 33 CISISRWETYTDDSNNWN 50
CIS+S W+T S NWN
Sbjct: 613 CISLSAWKTIVV*SKNWN 666
>TC82916 weakly similar to GP|20067231|gb|AAM09563.1 REP13 {Secale cereale},
partial (17%)
Length = 844
Score = 26.6 bits (57), Expect = 7.5
Identities = 21/70 (30%), Positives = 36/70 (51%)
Frame = -3
Query: 70 SSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIGLTE 129
+ PG+ L + V+ V++G +KDA + A D+ +S S +AA++ + K G +
Sbjct: 671 ADPGDELKQGARDVQKNVSRGAEDAKDA--VTDAGDKLKSRSDSAADEGEGMLKKAG--K 504
Query: 130 TINSGIETFK 139
IN E K
Sbjct: 503 NINRAAEDVK 474
>TC76781 similar to GP|12539609|gb|AAG59584.1 pathogen-inducible
alpha-dioxygenase {Nicotiana attenuata}, partial (94%)
Length = 2232
Score = 26.2 bits (56), Expect = 9.8
Identities = 20/67 (29%), Positives = 32/67 (46%)
Frame = +2
Query: 34 ISISRWETYTDDSNNWNNLTPNHEDSITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVL 93
I IS+WE TDD L + D + ++D V MA++ + KGF +
Sbjct: 1571 IPISKWEDLTDDKEVIKVLEEIYGDDV--EELDVLVG------LMAEKKI-----KGFAI 1711
Query: 94 SKDAFVM 100
S+ AF++
Sbjct: 1712 SETAFLI 1732
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.122 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,903,781
Number of Sequences: 36976
Number of extensions: 45849
Number of successful extensions: 219
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 204
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 205
length of query: 212
length of database: 9,014,727
effective HSP length: 92
effective length of query: 120
effective length of database: 5,612,935
effective search space: 673552200
effective search space used: 673552200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148762.7


