

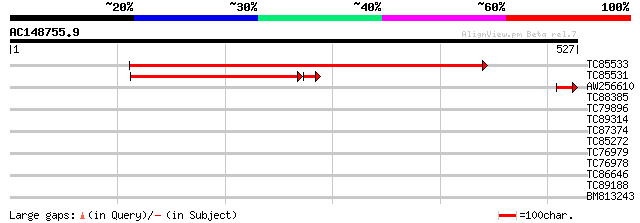
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.9 + phase: 0
(527 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85533 similar to SP|P42043|HMZ1_ARATH Ferrochelatase I chloro... 506 e-144
TC85531 similar to SP|P42044|HEMZ_CUCSA Ferrochelatase II chlor... 243 3e-68
AW256610 GP|15147828|em ferrochelatase {Nicotiana tabacum}, part... 45 5e-05
TC88385 similar to GP|22136512|gb|AAM91334.1 unknown protein {Ar... 35 0.054
TC79896 similar to GP|15795154|dbj|BAB03142. gene_id:MQC3.16~unk... 32 0.46
TC89314 similar to GP|21617911|gb|AAM66961.1 unknown {Arabidopsi... 32 0.46
TC87374 similar to PIR|T52310|T52310 Lil3 protein [imported] - A... 32 0.60
TC85272 similar to SP|Q02060|PSBS_SPIOL Photosystem II 22 kDa pr... 31 1.3
TC76979 homologue to GP|14487954|gb|AAK63815.1 early light induc... 30 2.3
TC76978 homologue to GP|14487954|gb|AAK63815.1 early light induc... 30 2.3
TC86646 similar to PIR|E84471|E84471 probable beta-1 3-glucanase... 29 5.1
TC89188 similar to PIR|S59541|S59541 heat shock transcription fa... 28 6.6
BM813243 homologue to PIR|B86439|B86 protein T19E23.11 [imported... 28 8.7
>TC85533 similar to SP|P42043|HMZ1_ARATH Ferrochelatase I
chloroplast/mitochondrial precursor (EC 4.99.1.1)
(Protoheme ferro-lyase), partial (76%)
Length = 1667
Score = 506 bits (1303), Expect = e-144
Identities = 244/333 (73%), Positives = 293/333 (87%)
Frame = +2
Query: 112 DDKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSK 171
++++GVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLF FLQ+PLA+ +S LRAPKSK
Sbjct: 263 EERVGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFRFLQQPLAKLISTLRAPKSK 442
Query: 172 EGYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGIT 231
E YASIGGGSPLR++TD QA L+++L K + +N+YVGMRYW+PFTEEAI+ IK+DGI
Sbjct: 443 EAYASIGGGSPLRKITDDQALALKRALEAKGLSSNIYVGMRYWYPFTEEAIQQIKKDGIR 622
Query: 232 KLVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEK 291
+LVVLPLYPQFSIST+GSS+ +LE FRED YL + ++I SWYQREGYIK+MA+LIEK
Sbjct: 623 RLVVLPLYPQFSISTTGSSISVLEQTFREDAYLSRLPVSIINSWYQREGYIKSMADLIEK 802
Query: 292 ELKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAY 351
EL+ F P++ MIFFSAHGVPV+YVE AGDPY+ +MEEC+ LIM+EL+ R I+N +TLAY
Sbjct: 803 ELESFSEPKEAMIFFSAHGVPVSYVENAGDPYRDQMEECIFLIMQELKARGISNEHTLAY 982
Query: 352 QSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI 411
QSRVGPV+WLKPYTDE ++ELG+KGVKSLLAVP+SFVSEHIETLEEID+EY+ELALESGI
Sbjct: 983 QSRVGPVQWLKPYTDEVLVELGQKGVKSLLAVPVSFVSEHIETLEEIDMEYRELALESGI 1162
Query: 412 ENWGRVPALGCEPTFISDLADAVIESLPYVGAM 444
+NW RVPALG P+FI DLADAVIE+LP A+
Sbjct: 1163KNWARVPALGLTPSFIMDLADAVIEALPSAAAI 1261
>TC85531 similar to SP|P42044|HEMZ_CUCSA Ferrochelatase II chloroplast
precursor (EC 4.99.1.1) (Protoheme ferro-lyase) (Heme
synthetase)., partial (34%)
Length = 995
Score = 243 bits (619), Expect(2) = 3e-68
Identities = 117/160 (73%), Positives = 140/160 (87%)
Frame = +2
Query: 113 DKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKE 172
+K+GVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLF FLQ+PLA+ +S LRAPKSKE
Sbjct: 464 EKVGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFRFLQQPLAKLISTLRAPKSKE 643
Query: 173 GYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITK 232
YASIGGGSPLR++TD QA L+++L K + +N+YVGMRYW+PFTEEAI+ IK+DGIT+
Sbjct: 644 AYASIGGGSPLRKITDDQALALKRALEAKGLSSNIYVGMRYWYPFTEEAIQQIKKDGITR 823
Query: 233 LVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVI 272
LVVLPLYPQFSIST+GSS+ +LE FRED YL + ++I
Sbjct: 824 LVVLPLYPQFSISTTGSSISVLEQTFREDAYLSRLPVSII 943
Score = 34.7 bits (78), Expect(2) = 3e-68
Identities = 14/16 (87%), Positives = 16/16 (99%)
Frame = +3
Query: 274 SWYQREGYIKAMANLI 289
SWYQREGYIK+MA+LI
Sbjct: 948 SWYQREGYIKSMADLI 995
>AW256610 GP|15147828|em ferrochelatase {Nicotiana tabacum}, partial (3%)
Length = 300
Score = 45.4 bits (106), Expect = 5e-05
Identities = 19/19 (100%), Positives = 19/19 (100%)
Frame = -2
Query: 509 EVTTGEGFLHQWGILPLFR 527
EVTTGEGFLHQWGILPLFR
Sbjct: 299 EVTTGEGFLHQWGILPLFR 243
>TC88385 similar to GP|22136512|gb|AAM91334.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 1001
Score = 35.4 bits (80), Expect = 0.054
Identities = 18/40 (45%), Positives = 25/40 (62%)
Frame = +2
Query: 484 WGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
+G+ + E NG+AA+I LLLL E+ TG+G L G L
Sbjct: 716 FGFNRKNEIINGKAAVIGFLLLLDFELLTGKGLLKGTGFL 835
>TC79896 similar to GP|15795154|dbj|BAB03142. gene_id:MQC3.16~unknown
protein {Arabidopsis thaliana}, partial (58%)
Length = 739
Score = 32.3 bits (72), Expect = 0.46
Identities = 13/36 (36%), Positives = 25/36 (69%)
Frame = +1
Query: 483 EWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLH 518
E G+++ +E NGR A++ + LL +E+ TG+G ++
Sbjct: 373 ELGFSRYSERLNGRIALLGLTALLLVELATGKGVIN 480
>TC89314 similar to GP|21617911|gb|AAM66961.1 unknown {Arabidopsis
thaliana}, partial (52%)
Length = 755
Score = 32.3 bits (72), Expect = 0.46
Identities = 11/32 (34%), Positives = 21/32 (65%)
Frame = +2
Query: 491 ETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGI 522
+ W GR AM+ + + +E+T+G+G L +G+
Sbjct: 416 DVWLGRVAMVGFAVAITVEITSGKGLLENFGL 511
>TC87374 similar to PIR|T52310|T52310 Lil3 protein [imported] - Arabidopsis
thaliana, partial (44%)
Length = 1119
Score = 32.0 bits (71), Expect = 0.60
Identities = 18/46 (39%), Positives = 23/46 (49%), Gaps = 5/46 (10%)
Frame = +2
Query: 479 ILVWEWGWTK-----SAETWNGRAAMIAVLLLLFLEVTTGEGFLHQ 519
I+ W W W K AE NGRAAMI + ++ TG G + Q
Sbjct: 644 IVPW-WAWIKRFHLPEAELLNGRAAMIGFFMTYLVDSLTGVGLVDQ 778
>TC85272 similar to SP|Q02060|PSBS_SPIOL Photosystem II 22 kDa protein
chloroplast precursor (CP22). [Spinach] {Spinacia
oleracea}, partial (76%)
Length = 970
Score = 30.8 bits (68), Expect = 1.3
Identities = 16/39 (41%), Positives = 23/39 (58%)
Frame = +3
Query: 484 WGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGI 522
+G+TKS E + GR A + + L E+ TG+G L Q I
Sbjct: 645 FGFTKSNELFVGRLAQLGFVFSLIGEIITGKGALAQLNI 761
Score = 29.3 bits (64), Expect = 3.9
Identities = 15/36 (41%), Positives = 20/36 (54%)
Frame = +3
Query: 484 WGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQ 519
+G+TK E + GR AMI + E TG+G L Q
Sbjct: 339 FGFTKQNELFVGRVAMIGFAASILGEALTGKGILAQ 446
>TC76979 homologue to GP|14487954|gb|AAK63815.1 early light inducible
protein {Medicago sativa}, partial (93%)
Length = 787
Score = 30.0 bits (66), Expect = 2.3
Identities = 12/27 (44%), Positives = 16/27 (58%)
Frame = +1
Query: 487 TKSAETWNGRAAMIAVLLLLFLEVTTG 513
+ AE WNGR AM+ ++ L F E G
Sbjct: 616 SSDAELWNGRIAMLGLIALAFTEYVKG 696
>TC76978 homologue to GP|14487954|gb|AAK63815.1 early light inducible
protein {Medicago sativa}, complete
Length = 953
Score = 30.0 bits (66), Expect = 2.3
Identities = 12/27 (44%), Positives = 16/27 (58%)
Frame = +2
Query: 487 TKSAETWNGRAAMIAVLLLLFLEVTTG 513
+ AE WNGR AM+ ++ L F E G
Sbjct: 626 SSDAELWNGRIAMLGLIALAFTEYVKG 706
>TC86646 similar to PIR|E84471|E84471 probable beta-1 3-glucanase [imported]
- Arabidopsis thaliana, partial (73%)
Length = 1257
Score = 28.9 bits (63), Expect = 5.1
Identities = 14/63 (22%), Positives = 28/63 (44%)
Frame = +2
Query: 216 PFTEEAIELIKRDGITKLVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSW 275
P + ++L+K GIT + + P + SGS ++L + + + + SW
Sbjct: 182 PSATKVVQLLKSQGITHVKIYDTDPSVLRALSGSKIKLTVDLPNQQLFAAAKSQSFALSW 361
Query: 276 YQR 278
+R
Sbjct: 362 VER 370
>TC89188 similar to PIR|S59541|S59541 heat shock transcription factor HSF29
- soybean (fragment), partial (72%)
Length = 1493
Score = 28.5 bits (62), Expect = 6.6
Identities = 14/31 (45%), Positives = 17/31 (54%)
Frame = -1
Query: 12 SCSYIRPHSCRTCASRNFKLPLLLPQAICTA 42
SCSY R H C C R + PL +P+ I A
Sbjct: 395 SCSYSRRHRCSCC--RQYFSPLYIPKQIFLA 309
>BM813243 homologue to PIR|B86439|B86 protein T19E23.11 [imported] -
Arabidopsis thaliana, partial (66%)
Length = 725
Score = 28.1 bits (61), Expect = 8.7
Identities = 13/36 (36%), Positives = 15/36 (41%)
Frame = +3
Query: 453 QSLVPLGSVEELLAAYDSQRRELPPPILVWEWGWTK 488
Q +V L L S R P L+W W WTK
Sbjct: 441 QDIVQLAQPIVALHHQKSMNRNKPKIFLIWTWWWTK 548
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,520,749
Number of Sequences: 36976
Number of extensions: 237848
Number of successful extensions: 1192
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1178
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1191
length of query: 527
length of database: 9,014,727
effective HSP length: 101
effective length of query: 426
effective length of database: 5,280,151
effective search space: 2249344326
effective search space used: 2249344326
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148755.9


