

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.7 - phase: 0
(503 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
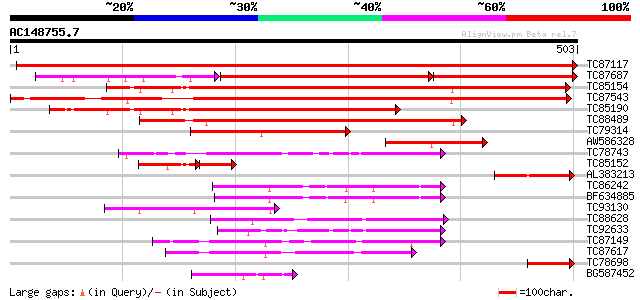
Score E
Sequences producing significant alignments: (bits) Value
TC87117 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 1002 0.0
TC87687 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 332 e-177
TC85154 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 534 e-152
TC87543 similar to PIR|A96745|A96745 probable cytosolic factor T... 474 e-134
TC85190 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 326 1e-89
TC88489 similar to GP|14334978|gb|AAK59666.1 unknown protein {Ar... 288 3e-78
TC79314 similar to GP|16930483|gb|AAL31927.1 AT3g51670/T18N14_50... 163 1e-40
AW586328 weakly similar to GP|20804751|dbj contains ESTs AU09391... 141 5e-34
TC78743 similar to PIR|T05953|T05953 polyphosphoinositide bindin... 92 5e-19
TC85152 similar to GP|8894548|emb|CAB95829.1 hypothetical protei... 63 8e-19
AL383213 weakly similar to GP|14334978|gb| unknown protein {Arab... 91 1e-18
TC86242 similar to GP|14486705|gb|AAK63247.1 phosphatidylinosito... 79 5e-15
BF634885 similar to GP|11994666|db phosphatidylinositol/phosphat... 70 2e-12
TC93130 weakly similar to GP|16930483|gb|AAL31927.1 AT3g51670/T1... 69 3e-12
TC88628 similar to PIR|B86282|B86282 protein F10B6.22 [imported]... 63 2e-10
TC92633 similar to PIR|T05953|T05953 polyphosphoinositide bindin... 62 7e-10
TC87149 weakly similar to PIR|D86354|D86354 hypothetical protein... 60 1e-09
TC87617 similar to PIR|D86354|D86354 hypothetical protein AAF878... 59 3e-09
TC78698 homologue to GP|3746903|gb|AAC64109.1| signal recognitio... 55 5e-08
BG587452 weakly similar to SP|Q10138|YAS2_ Hypothetical protein ... 52 5e-07
>TC87117 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (72%)
Length = 1662
Score = 1002 bits (2590), Expect = 0.0
Identities = 497/497 (100%), Positives = 497/497 (100%)
Frame = +3
Query: 7 SNQTTPHVYVPQPLYSHPQPQKKNNPVTLQSHLSKPNTEQEQTFNKPSDNIPNNENNSLQ 66
SNQTTPHVYVPQPLYSHPQPQKKNNPVTLQSHLSKPNTEQEQTFNKPSDNIPNNENNSLQ
Sbjct: 9 SNQTTPHVYVPQPLYSHPQPQKKNNPVTLQSHLSKPNTEQEQTFNKPSDNIPNNENNSLQ 188
Query: 67 ELQNLIQQAFNNHAFSAPPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGA 126
ELQNLIQQAFNNHAFSAPPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGA
Sbjct: 189 ELQNLIQQAFNNHAFSAPPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGA 368
Query: 127 KTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFL 186
KTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFL
Sbjct: 369 KTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFL 548
Query: 187 RARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYG 246
RARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYG
Sbjct: 549 RARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYG 728
Query: 247 EFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGK 306
EFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGK
Sbjct: 729 EFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGK 908
Query: 307 WELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSK 366
WELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSK
Sbjct: 909 WELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSK 1088
Query: 367 STETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEV 426
STETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEV
Sbjct: 1089STETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEV 1268
Query: 427 RVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSS 486
RVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSS
Sbjct: 1269RVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSS 1448
Query: 487 RKKKLLYRLKTKTSLSD 503
RKKKLLYRLKTKTSLSD
Sbjct: 1449RKKKLLYRLKTKTSLSD 1499
>TC87687 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (67%)
Length = 1969
Score = 332 bits (851), Expect(3) = e-177
Identities = 166/192 (86%), Positives = 171/192 (88%), Gaps = 3/192 (1%)
Frame = +1
Query: 188 ARDFKVKEAFTMIKNTILWRKEFGIEELM-DEKLGDE-LEKVVYMHGFDKEGHPVCYNIY 245
ARDFKVKEAFTMIKNTI WRKEF IEEL+ DE LGDE LEK VYMHG+DKEGHPVCYNIY
Sbjct: 766 ARDFKVKEAFTMIKNTIRWRKEFKIEELLLDENLGDEYLEKTVYMHGYDKEGHPVCYNIY 945
Query: 246 GEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPG 305
GEF NKE+Y KTFSDEEKR FLKWRI LEKSIR LDF GG+CTIV VNDLK+SPGP
Sbjct: 946 GEFDNKEVYKKTFSDEEKRERFLKWRILVLEKSIRKLDFTPGGICTIVQVNDLKNSPGPT 1125
Query: 306 KWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFA-GP 364
KWELRQATKQALQL QDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVF GP
Sbjct: 1126KWELRQATKQALQLLQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFCLGP 1305
Query: 365 SKSTETLLSYIA 376
SKS ETLLSYIA
Sbjct: 1306SKSAETLLSYIA 1341
Score = 217 bits (553), Expect(3) = e-177
Identities = 103/127 (81%), Positives = 117/127 (92%)
Frame = +2
Query: 377 PEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYG 436
PEQL VKYGGLSKDGEFG +D+VTEIT+RPA+KH VEFPVTE CLLSWE+RVIGW++ YG
Sbjct: 1343 PEQLSVKYGGLSKDGEFGITDAVTEITVRPAAKHIVEFPVTENCLLSWELRVIGWDISYG 1522
Query: 437 AEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLK 496
AEFVPS+EGSYTVI+QK RK+ASSEE VLCN++KI EPGKVVLTIDN+SS+KKKLLYRLK
Sbjct: 1523 AEFVPSSEGSYTVIIQKTRKIASSEEPVLCNNYKIGEPGKVVLTIDNSSSKKKKLLYRLK 1702
Query: 497 TKTSLSD 503
TK S SD
Sbjct: 1703 TKPSTSD 1723
Score = 111 bits (278), Expect(3) = e-177
Identities = 85/216 (39%), Positives = 108/216 (49%), Gaps = 53/216 (24%)
Frame = +3
Query: 24 PQPQKKNNPVTLQSHLSKPNTE----QEQTFNKPSD---NIPNNENNSLQELQNLIQQAF 76
P P+ N T +SKP+ + + +F + S +P E +L EL+ LIQ+A
Sbjct: 126 PNPEPVENNTTETEEVSKPSGDDKIPESGSFKEESTIVGELPEAEKKALAELKQLIQEAL 305
Query: 77 NNHAFSAPPL-----IKEQKQSTTTTVAAE------------------------------ 101
N H FSAP +KEQK T AE
Sbjct: 306 NKHEFSAPASTTPSPVKEQKPEPTPEAPAEETNKKDEQVSETESVVAVTTADDDVSTTPP 485
Query: 102 -PAQENKYQLEDKKENV--------VSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVE 152
P E + + E KE V SSV++DGAKTVEAIEES+VAV++SVP E K
Sbjct: 486 PPPTEAEAEAEQPKEEVEKKETEVAASSVDEDGAKTVEAIEESVVAVASSVPEEPK---- 653
Query: 153 KVEASLP--LPPEQVSIYGIPLLADETSDVILLKFL 186
VEAS P PE+VSI+GIPLLAD+ +DVILLKFL
Sbjct: 654 VVEASSPEQQQPEEVSIWGIPLLADDRTDVILLKFL 761
>TC85154 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (83%)
Length = 1811
Score = 534 bits (1375), Expect = e-152
Identities = 276/425 (64%), Positives = 331/425 (76%), Gaps = 14/425 (3%)
Frame = +3
Query: 87 IKEQKQSTTTTVAAEPAQENKYQLEDKK---------ENVVSSVEDDGAKTVEAIEESIV 137
+ E+K ST+ E A+ K E+KK E + +S E+DGAKTVEAI+ESIV
Sbjct: 186 VDEEKGSTSEEPKVETAEPEK---EEKKVEETVVEVVEKIAASTEEDGAKTVEAIQESIV 356
Query: 138 AVSASV--PPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKE 195
+V + P +PV E VE + P+ PE+V I+GIPLLADE SDVILLKFLRARDFKVKE
Sbjct: 357 SVPVTEGEQPVAEPVAE-VEVT-PIVPEEVEIWGIPLLADERSDVILLKFLRARDFKVKE 530
Query: 196 AFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYN 255
AFTMIK T+LWRKEFG+E L+ E LG + +KVV+ G DKEGHPV YN++GEF++K+LY
Sbjct: 531 AFTMIKQTVLWRKEFGVEALLQEDLGTDWDKVVFTDGTDKEGHPVYYNVFGEFEDKDLYQ 710
Query: 256 KTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPG-PGKWELRQATK 314
KTFSDEEKR F++W IQ LEKS+R LDF G+ T+V +NDLK+SPG GK ELRQ+ K
Sbjct: 711 KTFSDEEKRTKFVRWWIQSLEKSVRKLDFAPSGISTLVQINDLKNSPGLLGKKELRQSIK 890
Query: 315 QALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSY 374
Q LQL QDNYPEFVAKQ+FINVPWWYLA +RMISPFLTQRTKSKFVFAG SKS ETL Y
Sbjct: 891 QTLQLLQDNYPEFVAKQIFINVPWWYLAFSRMISPFLTQRTKSKFVFAGSSKSAETLFKY 1070
Query: 375 IAPEQLPVKYGGLSKDG--EFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWE 432
IAPEQ+PVKYGGLS+DG EF +D TE+TI+PA+KH VEFPV+EK L WEVRV+ W
Sbjct: 1071IAPEQVPVKYGGLSRDGEQEFTTADPATEVTIKPATKHAVEFPVSEKSTLVWEVRVVDWN 1250
Query: 433 VRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLL 492
V YGAEFVPS E YTVI+QK RK+A+++E V+ N+FK+ EPGKVVLTIDN +S+KKKLL
Sbjct: 1251VSYGAEFVPSAEDGYTVIIQKNRKIAAADETVISNTFKVGEPGKVVLTIDNQTSKKKKLL 1430
Query: 493 YRLKT 497
YR KT
Sbjct: 1431YRSKT 1445
>TC87543 similar to PIR|A96745|A96745 probable cytosolic factor T9N14.8
[imported] - Arabidopsis thaliana, partial (74%)
Length = 1718
Score = 474 bits (1220), Expect = e-134
Identities = 248/505 (49%), Positives = 325/505 (64%), Gaps = 7/505 (1%)
Frame = +1
Query: 1 MSHDDSSNQTTPHVYVPQPLYSHPQPQKKNNPVTLQSHLSKPNTEQEQTFNKPSDNIPNN 60
M+ ++ N T PH + YS P + PVT ++ L+ ++Q +T N
Sbjct: 97 MAENNPPNPTPPH----EETYSSPLTPEPELPVTQKTELTDEPSQQPETVTDTKGNPSQE 264
Query: 61 ENNSLQELQNLIQQAFNNHAFSAPPLIKEQKQSTTTTVAAEPA--QENKYQLEDKKENVV 118
E+ E S P +EQK+ + +E ++ D E+
Sbjct: 265 EDVVAAE-------------DSTEPSKEEQKEPNQNKIPQNLGSFKEESNRVTDLSESER 405
Query: 119 SSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETS 178
+S++ +++++ +QVSIYG+PLL DE +
Sbjct: 406 TSLQQFKTLLTDSLKDD---------------------------QQVSIYGVPLLEDERT 504
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGH 238
D ILLKFLRARDFK KE+ TM+KNT+ WRK F I+ L+DE LGD+L+KVV+MHGF +EGH
Sbjct: 505 DTILLKFLRARDFKPKESHTMLKNTLQWRKSFNIDALLDEDLGDDLDKVVFMHGFSREGH 684
Query: 239 PVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDL 298
PVCYN+YGEFQNKELY KTF EEKR FL+WR+QFLEKSIR LDF+ GGV T+ VNDL
Sbjct: 685 PVCYNVYGEFQNKELYEKTFGSEEKRERFLRWRVQFLEKSIRKLDFSPGGVNTLFQVNDL 864
Query: 299 KDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSK 358
K+SPGP K ELR ATK AL+L QDNYPEFVAKQVFINVPWWYLA +++PFLTQRTKSK
Sbjct: 865 KNSPGPAKKELRVATKMALELLQDNYPEFVAKQVFINVPWWYLAFYTILNPFLTQRTKSK 1044
Query: 359 FVFAGPSKSTETLLSYIAPEQLPVKYGGLSKD-----GEFGNSDSVTEITIRPASKHTVE 413
FVFAG SKS +TL YI PEQ+PV+YGGLS D +F +D TEI ++P++K TVE
Sbjct: 1045FVFAGTSKSPDTLFKYITPEQVPVQYGGLSVDFCDCNPDFSINDPTTEIPVKPSTKQTVE 1224
Query: 414 FPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINE 473
+ EKC++ WE+RV+GWEV Y AEF P ++ +Y VI+QKA K+ ++E V+ NSFK+ E
Sbjct: 1225IAIYEKCIIVWELRVVGWEVSYSAEFKPDDKDAYGVIIQKATKMTPTDEPVVSNSFKVAE 1404
Query: 474 PGKVVLTIDNTSSRKKKLLYRLKTK 498
GK+ LT+DN + +KK+LLYR K K
Sbjct: 1405LGKLFLTVDNPTVKKKRLLYRFKIK 1479
>TC85190 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (50%)
Length = 1496
Score = 326 bits (836), Expect = 1e-89
Identities = 187/365 (51%), Positives = 238/365 (64%), Gaps = 54/365 (14%)
Frame = +3
Query: 36 QSHLSKPNTEQEQTFNKPSDNIPNNENNSLQELQNLIQQAFNNHAFSAPP---------- 85
+S +S+ + +E+T N S+ +P+ + +L EL+ LIQ+A N H F+APP
Sbjct: 369 ESKVSQSVSFKEET-NVVSE-LPDVQKKALDELKQLIQEALNKHEFTAPPPAPVKAPEPE 542
Query: 86 --------------------------------LIKEQKQSTTTTVAAEPAQENKYQLEDK 113
+ E+K ST+ E A+ K E+K
Sbjct: 543 VAVKEEKKPEEDEKKTEEVVEEKKDEAVVEEKKVDEEKGSTSEEPKVETAEPEK---EEK 713
Query: 114 K---------ENVVSSVEDDGAKTVEAIEESIVAVSASV--PPEQKPVVEKVEASLPLPP 162
K E + +S E+DGAKTVEAI+ESIV+V + P +PV E VE + P+ P
Sbjct: 714 KVEETVVEVVEKIAASTEEDGAKTVEAIQESIVSVPVTEGEQPVAEPVAE-VEVT-PIVP 887
Query: 163 EQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGD 222
E+V I+GIPLLADE SDVILLKFLRARDFKVKEAFTMIK T+LWRKEFG+E L+ E LG
Sbjct: 888 EEVEIWGIPLLADERSDVILLKFLRARDFKVKEAFTMIKQTVLWRKEFGVEALLQEDLGT 1067
Query: 223 ELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNL 282
+ +KVV+ G DKEGHPV YN++GEF++K+LY KTFSDEEKR F++W IQ LEKS+R L
Sbjct: 1068DWDKVVFTDGTDKEGHPVYYNVFGEFEDKDLYQKTFSDEEKRTKFVRWWIQSLEKSVRKL 1247
Query: 283 DFNHGGVCTIVHVNDLKDSPG-PGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYL 341
DF G+ T+V +NDLK+SPG GK ELRQ+ KQ LQL QDNYPEFVAKQ+FINVPWWYL
Sbjct: 1248DFAPSGISTLVQINDLKNSPGLLGKKELRQSIKQTLQLLQDNYPEFVAKQIFINVPWWYL 1427
Query: 342 AVNRM 346
A +RM
Sbjct: 1428AFSRM 1442
>TC88489 similar to GP|14334978|gb|AAK59666.1 unknown protein {Arabidopsis
thaliana}, partial (47%)
Length = 1187
Score = 288 bits (738), Expect = 3e-78
Identities = 146/297 (49%), Positives = 200/297 (67%), Gaps = 7/297 (2%)
Frame = +3
Query: 116 NVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPV-VEKVEASLPLPPEQVSIYGIPLL- 173
N + +++ K VE + E S V E++ VE+VE + +S++G+ LL
Sbjct: 312 NTLFEKKEEETKKVETLPEGGEESSEKVVKEEEEKRVEEVEE------KDLSLWGVSLLP 473
Query: 174 --ADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMH 231
+E DV+LLKFLRAR+FKV EAF M+K T+ WRKE I+ +++E G +L YM+
Sbjct: 474 SKGNEGVDVVLLKFLRAREFKVNEAFEMLKKTLKWRKEMKIDSVLEEDFGSDLASAAYMN 653
Query: 232 GFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCT 291
G D+EGHPVCYNIY F +E+Y KTF EEKR FL+WR +EK I+ L+ GGV +
Sbjct: 654 GVDREGHPVCYNIYSVFDGEEIYQKTFGTEEKRKEFLRWRCSVMEKWIQKLNLKPGGVSS 833
Query: 292 IVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFL 351
++ +NDLK+SPGP K ELR ATKQ + + QDNYPE VAK +FINVP+WY A+N ++SPFL
Sbjct: 834 LLQINDLKNSPGPSKKELRIATKQTVTMLQDNYPELVAKNIFINVPFWYYALNALLSPFL 1013
Query: 352 TQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGE---FGNSDSVTEITIR 405
TQRTKSKFV A P+K TETL+ YI E++PV YGG ++ + FG SV+E+ ++
Sbjct: 1014TQRTKSKFVVARPAKVTETLIKYIPIEEIPVNYGGFKRENDSEFFGQDASVSELFLK 1184
>TC79314 similar to GP|16930483|gb|AAL31927.1 AT3g51670/T18N14_50
{Arabidopsis thaliana}, partial (33%)
Length = 868
Score = 163 bits (413), Expect = 1e-40
Identities = 81/146 (55%), Positives = 105/146 (71%), Gaps = 4/146 (2%)
Frame = +1
Query: 161 PPEQVSIYGIPLLA-DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEK 219
P +VS++GIPLL D+ +DVILLKFLRARDF+V EA MI + WRKEF + +++E
Sbjct: 430 PNGEVSMWGIPLLGGDDRADVILLKFLRARDFRVNEALNMITKCLAWRKEFNADTIVEED 609
Query: 220 LGD--ELEKVV-YMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLE 276
L ELE V+ YM G+DKEGHPVCYN YG F++K++Y + F DEEK FL+WR+Q LE
Sbjct: 610 LSQFKELEGVIAYMQGYDKEGHPVCYNAYGLFRDKDMYERIFGDEEKLKKFLRWRVQVLE 789
Query: 277 KSIRNLDFNHGGVCTIVHVNDLKDSP 302
+ IR L F GGV +++ V DLKD P
Sbjct: 790 RGIRLLHFKPGGVNSLIQVTDLKDMP 867
>AW586328 weakly similar to GP|20804751|dbj contains ESTs AU093915(E1276)
AU162319(E60301)~unknown protein, partial (11%)
Length = 447
Score = 141 bits (356), Expect = 5e-34
Identities = 76/118 (64%), Positives = 84/118 (70%), Gaps = 27/118 (22%)
Frame = +1
Query: 334 INVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLL--------------------- 372
INVPW YL+VNRMISPFLTQRTKSKFVFAGPSKS ETLL
Sbjct: 1 INVPWXYLSVNRMISPFLTQRTKSKFVFAGPSKSAETLLR*RKKHISCSCLFSLVC*CLS 180
Query: 373 ------SYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSW 424
SYIAPEQL VKYGGLSKDGEFG +D+VTEIT+RPA+KH VEFPVTE L+++
Sbjct: 181 MFYGFFSYIAPEQLSVKYGGLSKDGEFGITDAVTEITVRPAAKHIVEFPVTEVSLVAF 354
>TC78743 similar to PIR|T05953|T05953 polyphosphoinositide binding protein
Ssh2 - soybean, partial (94%)
Length = 1232
Score = 92.0 bits (227), Expect = 5e-19
Identities = 85/296 (28%), Positives = 133/296 (44%), Gaps = 6/296 (2%)
Frame = +1
Query: 97 TVAAEP----AQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVE 152
TV EP A ++K++L ++ V+DDGA ++ E + ++ + +VE
Sbjct: 205 TVNPEPGRNSALDSKHELAKEETKGKILVQDDGALN-DSTEAELTKINL-----MRTLVE 366
Query: 153 KVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGI 212
+ S + E D+++ +FLRARD V +A M + WRK F
Sbjct: 367 SRDPS----------------SKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVP 498
Query: 213 E-ELMDEKLGDEL-EKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKW 270
+ ++ D+L ++ +Y+ G DK+G P+ F K NK D KR+
Sbjct: 499 SGSVSPSEIADDLAQEKIYVQGLDKKGRPIIV----AFAAKHFQNKNGLDAFKRYVVFA- 663
Query: 271 RIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAK 330
LEK I + G V + D+K G ++R AL + QD YPE + K
Sbjct: 664 ----LEKLISRMP---PGEEKFVSIADIKGW-GYANSDIR-GYLGALTILQDYYPERLGK 816
Query: 331 QVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
++ P+ ++ V ++I PF+ TK K VF K TLL I QLP YGG
Sbjct: 817 LFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 984
>TC85152 similar to GP|8894548|emb|CAB95829.1 hypothetical protein {Cicer
arietinum}, partial (18%)
Length = 775
Score = 62.8 bits (151), Expect(2) = 8e-19
Identities = 31/33 (93%), Positives = 32/33 (96%)
Frame = +1
Query: 169 GIPLLADETSDVILLKFLRARDFKVKEAFTMIK 201
GIPLLADE SDVILLKFLRA+DFKVKEAFTMIK
Sbjct: 676 GIPLLADERSDVILLKFLRAKDFKVKEAFTMIK 774
Score = 48.9 bits (115), Expect(2) = 8e-19
Identities = 29/57 (50%), Positives = 40/57 (69%), Gaps = 2/57 (3%)
Frame = +3
Query: 115 ENVVSSVEDDGAKTVEAIEESIVA--VSASVPPEQKPVVEKVEASLPLPPEQVSIYG 169
E + +S E+DGAKTVEAI+ESIV+ V+ P +PV E VE + P+ PE+V I+G
Sbjct: 513 EKIAASTEEDGAKTVEAIQESIVSVPVTEGEQPVAEPVAE-VEVT-PIVPEEVEIWG 677
>AL383213 weakly similar to GP|14334978|gb| unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 437
Score = 90.5 bits (223), Expect = 1e-18
Identities = 43/71 (60%), Positives = 58/71 (81%)
Frame = -2
Query: 431 WEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKK 490
WEV Y EFVP+++GSYTVIVQK +K+ S +E + N+FK NEPGKV+LTI+N+S++KK+
Sbjct: 436 WEVSYKEEFVPNDDGSYTVIVQKGKKIGS-QEGPIRNTFKNNEPGKVILTINNSSNKKKR 260
Query: 491 LLYRLKTKTSL 501
+LYR KT SL
Sbjct: 259 VLYRYKTNKSL 227
>TC86242 similar to GP|14486705|gb|AAK63247.1 phosphatidylinositol
transfer-like protein III {Lotus japonicus}, partial
(79%)
Length = 2271
Score = 78.6 bits (192), Expect = 5e-15
Identities = 62/218 (28%), Positives = 100/218 (45%), Gaps = 12/218 (5%)
Frame = +2
Query: 181 ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY-----MHGFDK 235
+LL+FL+AR F +++A M N I WRKE+G + +M++ EL++V+ HG DK
Sbjct: 485 MLLRFLKARKFDIEKAKLMWANMIQWRKEYGTDTIMEDFEFSELDEVLQYYPHGYHGVDK 664
Query: 236 EGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHV 295
EG PV G+ +L T + +LK+ +Q EK+ + F + H+
Sbjct: 665 EGRPVYIERLGKVDPNKLMQVTTME-----RYLKYHVQGFEKTFA-VKFPACSIAAKRHI 826
Query: 296 ND---LKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPWWYLAVNRMIS 348
+ + D G G + ++ +Q Q D YPE + + IN + + +
Sbjct: 827 DSSTTILDVHGVGFKNFSKIARELIQRLQKIDGDYYPETLCRMFIINAGPGFKLLWNTVK 1006
Query: 349 PFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
FL +T SK G +K L I +LP GG
Sbjct: 1007TFLDPKTSSKINVLG-NKFQSKLFEIIDVSELPEFLGG 1117
>BF634885 similar to GP|11994666|db phosphatidylinositol/phosphatidylcholine
transfer protein-like {Arabidopsis thaliana}, partial
(35%)
Length = 684
Score = 70.1 bits (170), Expect = 2e-12
Identities = 60/217 (27%), Positives = 94/217 (42%), Gaps = 12/217 (5%)
Frame = +3
Query: 182 LLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY-----MHGFDKE 236
+L+FL+ R F + + M + + WRKE+G + ++ + + E E+V MHG DKE
Sbjct: 27 MLRFLKGRKFDLDKTVQMWADMLHWRKEYGADSILQDFVYKEFEEVQRYYPHGMHGVDKE 206
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVN 296
G PV G+ +L N T D FL++ +Q EK + F + H++
Sbjct: 207 GRPVYIERLGKVDPIKLMNVTTID-----RFLRYHVQGFEKLFQE-KFPACSIAAKRHID 368
Query: 297 ---DLKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPWWYLAVNRMISP 349
+ D G + + + Q DNYPE + + +N + V
Sbjct: 369 KTTTIIDVHGVNWLSFSKIAHELVMRMQKIDGDNYPETLNQMFIVNAGSGFKLVWNTAKG 548
Query: 350 FLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
FL +T +K + G SK LL I QLP GG
Sbjct: 549 FLDPKTTAKIIVLG-SKFQSRLLEVIDSCQLPDFLGG 656
>TC93130 weakly similar to GP|16930483|gb|AAL31927.1 AT3g51670/T18N14_50
{Arabidopsis thaliana}, partial (16%)
Length = 661
Score = 69.3 bits (168), Expect = 3e-12
Identities = 49/163 (30%), Positives = 87/163 (53%), Gaps = 8/163 (4%)
Frame = +2
Query: 85 PLIKEQKQSTTTTVAAEPAQENKYQLEDK--KENVVSSVEDDGAKTVEAIEESIVAVSAS 142
P ++E+ S++ ++ + P E + K K +V+++ + T + +E +S
Sbjct: 134 PSMEEEHSSSSMSILSTPFLETPRKTPSKPNKRTIVTTLMEAATFTSSSFKEDTYFISNL 313
Query: 143 VPPEQKPVVEKVEASLPLPP--EQVSIYGIPLL-ADETSDVILLKFLRARDFKVKEAFTM 199
E+K + + E L S++G+ L+ D+ +DV+LLKFLRARDF+V +A+TM
Sbjct: 314 KSSEKKALKQLKEKLLASDEITNNGSMWGVCLIKGDDVADVLLLKFLRARDFRVNDAYTM 493
Query: 200 -IKNTILWRKEFGIEELMDEKLGDELEKVVYM--HGFDKEGHP 239
+K L K ++ L + + KV+ + HGFD+EGHP
Sbjct: 494 LVKYVFLGEKNLVLKMLWMKIWDSKSLKVLLLSPHGFDREGHP 622
>TC88628 similar to PIR|B86282|B86282 protein F10B6.22 [imported] -
Arabidopsis thaliana, partial (27%)
Length = 1087
Score = 63.2 bits (152), Expect = 2e-10
Identities = 55/216 (25%), Positives = 97/216 (44%), Gaps = 5/216 (2%)
Frame = +1
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEE--LMDEKLGDELE-KVVYMHGFDK 235
D L++FL AR +A M WR + + D ++ DELE + +++ G K
Sbjct: 184 DPTLMRFLIARSMDSDKAAKMFVQWQKWRATMVPNDGFISDSEVPDELETRKIFLQGLSK 363
Query: 236 EGHPVCY-NIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRN-LDFNHGGVCTIV 293
+ +PV F +K+ + F K+ + L+K+I + G ++
Sbjct: 364 DKYPVMIVQASRHFPSKD-----------QIQFKKFIVHLLDKTIASAFKGREVGNEKLI 510
Query: 294 HVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQ 353
V DL+ T Q Q YPE +AK +++PW++++V R +S FL +
Sbjct: 511 GVLDLQGISYKNVDARGLIT--GFQFLQSYYPECLAKCYILHMPWFFVSVWRFVSGFLDK 684
Query: 354 RTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSK 389
T+ K V + + +S + + LP +YGG +K
Sbjct: 685 ATQEKIVIISNEEEKKLFVSEVGEDILPEEYGGRAK 792
>TC92633 similar to PIR|T05953|T05953 polyphosphoinositide binding protein
Ssh2 - soybean, partial (89%)
Length = 806
Score = 61.6 bits (148), Expect = 7e-10
Identities = 54/208 (25%), Positives = 92/208 (43%), Gaps = 6/208 (2%)
Frame = +1
Query: 185 FLRARDFKVKEAFTMIKNTILWRKEF------GIEELMDEKLGDELEKVVYMHGFDKEGH 238
F RD V++A M + WR F + ++ ++ D+ + G DK G
Sbjct: 157 FYVPRDLDVEKASAMFLKYLKWRHSFVPNGSISLSQVPNQIAQDK----AFAQGHDKIGR 324
Query: 239 PVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDL 298
P+ ++G + F ++ F ++ + L+K ++ G V + +L
Sbjct: 325 PILL-VFG--------GRHFQKKDGLDEFKRFAVYILDKLCASMP---PGEEKFVGIAEL 468
Query: 299 KDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSK 358
K G +LR AL + QD YPE + K ++ P+ ++ V +++ PF+ +TK K
Sbjct: 469 KGW-GYSNSDLRGYIG-ALSILQDYYPERLGKLFILHAPYIFMKVWKIVYPFIDNKTKKK 642
Query: 359 FVFAGPSKSTETLLSYIAPEQLPVKYGG 386
VF +K TLL I QLP Y G
Sbjct: 643 IVFVENNKLKSTLLXDIDESQLPEIYXG 726
>TC87149 weakly similar to PIR|D86354|D86354 hypothetical protein AAF87855.1
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1540
Score = 60.5 bits (145), Expect = 1e-09
Identities = 61/264 (23%), Positives = 116/264 (43%), Gaps = 4/264 (1%)
Frame = +1
Query: 127 KTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFL 186
KT E ++AVS S EQ+ + +V + ++ S+Y SD + ++L
Sbjct: 124 KTASDGHEKMLAVSLS--QEQQAKIIEVRKLIGTLSDKESVY--------CSDASISRYL 273
Query: 187 RARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELE--KVVYMHGFDKEGHPVCYNI 244
+++++ VK+A M+K ++ WR+E+ EE+ + + E E K+ + K+G PV
Sbjct: 274 KSQNWNVKKASQMLKQSLKWRQEYKPEEITWDDVAKEAETGKMYRPNYCAKDGRPVLI-- 447
Query: 245 YGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGP 304
+ +KT +E +K + +E +I NL N V +V + S
Sbjct: 448 ---MRTNRQKSKTLVEE------IKHFVYCMENAILNLPPNQEQVIWLVDFHGFSLSSVS 600
Query: 305 GKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRT--KSKFVFA 362
K T++ + Q YP+ + + + P + M+ L + K KFV++
Sbjct: 601 FKM-----TREVSHILQKYYPQRLGLAIMYDAPGIFQPFFSMVKVLLETESYNKIKFVYS 765
Query: 363 GPSKSTETLLSYIAPEQLPVKYGG 386
+ +T+ +QL +GG
Sbjct: 766 NDQNTKKTMEGLFDMDQLEPAFGG 837
>TC87617 similar to PIR|D86354|D86354 hypothetical protein AAF87855.1
[imported] - Arabidopsis thaliana, partial (77%)
Length = 1222
Score = 59.3 bits (142), Expect = 3e-09
Identities = 52/225 (23%), Positives = 94/225 (41%), Gaps = 2/225 (0%)
Frame = +2
Query: 139 VSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFT 198
V +P EQ+ + +V+ + + +Y SD + ++LRAR + VK+A
Sbjct: 176 VKMLIPEEQQGKINEVKKLIGPLSGKALVY--------CSDASISRYLRARSWNVKKAAK 331
Query: 199 MIKNTILWRKEFGIEELMDEKLGDELE--KVVYMHGFDKEGHPVCYNIYGEFQNKELYNK 256
M+K T+ WR E+ EE+ E + +E E K+ + DK G V +
Sbjct: 332 MLKQTLKWRAEYKPEEIRWEDVAEEAETGKIYRSNYTDKHGRTVL-----------VMRP 478
Query: 257 TFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQA 316
+ + +K+ + +E +I NL + +V S K T++
Sbjct: 479 ARQNSKTTKGQIKYLVYCMENAILNLSPEQEQMVWLVDFQGFNMSHISIK-----VTRET 643
Query: 317 LQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVF 361
+ Q++YPE + + N P + M+ P L +T +K F
Sbjct: 644 AHVLQEHYPERLGLAILYNPPKIFEPFFTMVKPILDTKTYNKVKF 778
>TC78698 homologue to GP|3746903|gb|AAC64109.1| signal recognition particle
54 kDa subunit precursor {Pisum sativum}, partial (45%)
Length = 1090
Score = 55.5 bits (132), Expect = 5e-08
Identities = 25/42 (59%), Positives = 35/42 (82%)
Frame = -2
Query: 460 SEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTKTSL 501
S+E + N+FK NEPGKV+LTI+N+S++KK++LYR KT SL
Sbjct: 234 SQEGPIRNTFKNNEPGKVILTINNSSNKKKRVLYRYKTNKSL 109
>BG587452 weakly similar to SP|Q10138|YAS2_ Hypothetical protein C3H8.02 in
chromosome I. [Fission yeast] {Schizosaccharomyces
pombe}, partial (6%)
Length = 688
Score = 52.0 bits (123), Expect = 5e-07
Identities = 36/111 (32%), Positives = 55/111 (49%), Gaps = 17/111 (15%)
Frame = +1
Query: 162 PEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILW-----------RKEF 210
PE++ L + + D+ILL+FLRAR ++V+ A M+ +T+ W + E
Sbjct: 280 PEEIRQALWSLTSKDDPDMILLRFLRARKWEVENAIAMLLSTVKWLLQNNINDIIKKGEE 459
Query: 211 GIEELMDEKLGDE------LEKVVYMHGFDKEGHPVCYNIYGEFQNKELYN 255
G+E+ EK GD+ Y+ G D EG P+CY I F K+ N
Sbjct: 460 GMEKYFKEK-GDKGFMKQFTSGKSYLRGTDNEGRPICY-INARFHKKDEQN 606
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,142,152
Number of Sequences: 36976
Number of extensions: 217496
Number of successful extensions: 1166
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 1135
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1145
length of query: 503
length of database: 9,014,727
effective HSP length: 100
effective length of query: 403
effective length of database: 5,317,127
effective search space: 2142802181
effective search space used: 2142802181
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148755.7


