

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.1 + phase: 0
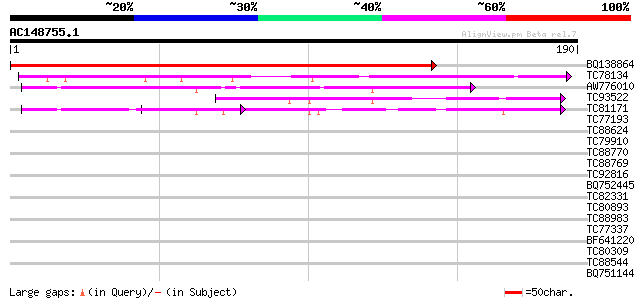
(190 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ138864 similar to GP|21553691|gb| unknown {Arabidopsis thalian... 290 2e-79
TC78134 similar to GP|9795609|gb|AAF98427.1| Unknown protein {Ar... 73 6e-14
AW776010 similar to GP|13877535|gb Unknown protein {Arabidopsis ... 60 5e-10
TC93522 weakly similar to PIR|C96836|C96836 unknown protein T21F... 54 5e-08
TC81171 similar to GP|9795609|gb|AAF98427.1| Unknown protein {Ar... 53 6e-08
TC77193 homologue to GP|21593495|gb|AAM65462.1 glycine rich prot... 39 0.002
TC88624 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 35 0.013
TC79910 similar to GP|9293979|dbj|BAB01882.1 gene_id:MAL21.25~un... 35 0.023
TC88770 homologue to GP|22202731|dbj|BAC07388. putative Rad6 {Or... 33 0.087
TC88769 homologue to GP|19881561|gb|AAM00962.1 hypothetical prot... 33 0.087
TC92816 weakly similar to GP|18076679|emb|CAC84774. P70 protein ... 33 0.087
BQ752445 similar to GP|18376387|em putative protein {Neurospora ... 32 0.19
TC82331 weakly similar to EGAD|120604|128852 hypothetical protei... 32 0.19
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 32 0.19
TC88983 similar to SP|P93231|VP41_LYCES Vacuolar assembly protei... 32 0.19
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 31 0.25
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 31 0.25
TC80309 similar to GP|20804963|dbj|BAB92640. hypothetical protei... 31 0.33
TC88544 similar to PIR|D96838|D96838 unknown protein T21F11.5 [i... 31 0.33
BQ751144 weakly similar to PIR|PQ0479|PQ04 pistil extensin-like ... 30 0.43
>BQ138864 similar to GP|21553691|gb| unknown {Arabidopsis thaliana}, partial
(23%)
Length = 668
Score = 290 bits (743), Expect = 2e-79
Identities = 143/143 (100%), Positives = 143/143 (100%)
Frame = +3
Query: 1 MEKPPVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTP 60
MEKPPVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTP
Sbjct: 216 MEKPPVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTP 395
Query: 61 PKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTP 120
PKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTP
Sbjct: 396 PKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTP 575
Query: 121 STLFSRLTIMEDEKKQGSVINEI 143
STLFSRLTIMEDEKKQGSVINEI
Sbjct: 576 STLFSRLTIMEDEKKQGSVINEI 644
>TC78134 similar to GP|9795609|gb|AAF98427.1| Unknown protein {Arabidopsis
thaliana}, partial (46%)
Length = 1133
Score = 73.2 bits (178), Expect = 6e-14
Identities = 71/213 (33%), Positives = 97/213 (45%), Gaps = 28/213 (13%)
Frame = +1
Query: 4 PPVHGSTT--GDCKPL-TTFVHTNTGAFREVVQRLTGPSEGNAT---------------- 44
PP+ T D P TTFV +T F++VVQ LTG SE +T
Sbjct: 256 PPLTPKTIPRSDSNPYPTTFVQADTTTFKQVVQMLTGSSETTSTTPTTTKPTTKPNQQQD 435
Query: 45 --KEEQTAKVAPIT---KRTPPKLHERRKCMKPK---LEIVKPNLQYHKQPGASPSSKSR 96
++ + + PI K+ KL+ERR +K L + PN S+
Sbjct: 436 LPQQTRNFNIPPIKTAPKKQGFKLYERRNSLKNSVLMLNTLMPNF-------------SQ 576
Query: 97 NSSF-PSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKER 155
NS+F P S SLL P S S +T + D+ S + N EE+KAI ER
Sbjct: 577 NSNFSPRKQQQEILSPSLLDFP---SLALSPVTPLNDDPFDKSSPSLGNSSEEDKAIAER 747
Query: 156 RFYLHPSPRSKASGFNEPELLNLFPLASPKACE 188
+YLHPSP + EP+LL LFPL+SP+ +
Sbjct: 748 GYYLHPSPITTPRD-AEPQLLPLFPLSSPRVSQ 843
>AW776010 similar to GP|13877535|gb Unknown protein {Arabidopsis thaliana},
partial (42%)
Length = 622
Score = 60.1 bits (144), Expect = 5e-10
Identities = 55/159 (34%), Positives = 75/159 (46%), Gaps = 7/159 (4%)
Frame = +2
Query: 5 PVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPP--- 61
PV S G+ P TTFV + +F++VVQ LTG +E + T+ + PP
Sbjct: 158 PVTRSEPGNPYP-TTFVQADPSSFKQVVQMLTGSTETTTKQTSSTSSNNNLRNHIPPKKS 334
Query: 62 --KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTT 119
KL+ERR +K L + P L +PSS S SP S +L+ SP T
Sbjct: 335 GFKLYERRNSLK-NLHL-NPLLPVFSSNSNNPSSFSPRKPEILSP-SILDFPALVLSPVT 505
Query: 120 P--STLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERR 156
P F+R T+ GS +NIE EEKA KE++
Sbjct: 506 PLIPDPFNRTTVSGHRCGNGSPKPGLNIEAEEKAYKEKK 622
>TC93522 weakly similar to PIR|C96836|C96836 unknown protein T21F11.22
[imported] - Arabidopsis thaliana, partial (20%)
Length = 599
Score = 53.5 bits (127), Expect = 5e-08
Identities = 42/120 (35%), Positives = 62/120 (51%), Gaps = 3/120 (2%)
Frame = +3
Query: 70 MKPKLEIVKPNLQYHKQPGASPS-SKSRNSS-FPSSPVSGSGSSSLLPSPTTP-STLFSR 126
M+P + + KPN + H++ + S N F ++ + + S ++ SP +P +S
Sbjct: 51 MEPPMGLKKPNFKLHERRSKKLQLNVSENPMMFQNNNNNNNMRSVVMASPVSPLEYYYSP 230
Query: 127 LTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPLASPKA 186
+ E+E+K EEEK I E+ FYLHPSPR AS P LL LFPL SP +
Sbjct: 231 KSPHENEEK-----------EEEKVIAEKGFYLHPSPR--ASQQQPPALLPLFPLHSPSS 371
>TC81171 similar to GP|9795609|gb|AAF98427.1| Unknown protein {Arabidopsis
thaliana}, partial (40%)
Length = 932
Score = 53.1 bits (126), Expect = 6e-08
Identities = 50/152 (32%), Positives = 72/152 (46%), Gaps = 10/152 (6%)
Frame = +1
Query: 45 KEEQTAKVAPITKRTPP-KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSS---- 99
+++QT + + K+ KL+ERR L I SP +K S
Sbjct: 346 QQKQTITITFLQKKQQGFKLYERRNSFHKNLNINPLLPPIFSNSTFSPRNKQEILSPSIL 525
Query: 100 -FPS---SPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKER 155
FPS SPV+ L+P P S S + GS ++ + E+KAI+E+
Sbjct: 526 DFPSLVLSPVT-----PLIPDPFNRSGSSSS----SSAARNGSSLDSL---AEDKAIREK 669
Query: 156 RFYLHPSPR-SKASGFNEPELLNLFPLASPKA 186
F+LHPSPR + S +EP LL LFP +SP+A
Sbjct: 670 GFFLHPSPRAASTSRDSEPRLLPLFPTSSPRA 765
Score = 40.0 bits (92), Expect = 5e-04
Identities = 30/76 (39%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Frame = +3
Query: 5 PVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLH 64
P+ S G+ P TTFV +T +F++VVQ LTG SE K+ T+ A PPK
Sbjct: 219 PITRSEPGNPYP-TTFVQADTTSFKQVVQMLTGSSE--TAKQASTSTKANHNHNIPPKKT 389
Query: 65 ERRKCM-KPKLEIVKP 79
R + + K KL KP
Sbjct: 390 TRFQTL*KKKLISQKP 437
>TC77193 homologue to GP|21593495|gb|AAM65462.1 glycine rich protein-like
{Arabidopsis thaliana}, partial (85%)
Length = 989
Score = 38.5 bits (88), Expect = 0.002
Identities = 33/112 (29%), Positives = 49/112 (43%), Gaps = 7/112 (6%)
Frame = -2
Query: 17 LTTFVHTNTGAFREV--VQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCM---- 70
L ++ +T F + ++ + T E + K A T + ++ KC+
Sbjct: 766 LPQIIYNSTNIFETID*YKKNASKLKSKETYETRAVKSAAGTHK------DKHKCL*K*S 605
Query: 71 KPKLEIVKPNLQYHKQPGASPS-SKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
K LE+V + H+ SPS S S S +SP S SGSSS TTPS
Sbjct: 604 KVPLEVVHTSYHLHRSTSTSPSTSASSRLSTLASPASASGSSSTTIIKTTPS 449
>TC88624 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(50%)
Length = 1496
Score = 35.4 bits (80), Expect = 0.013
Identities = 28/89 (31%), Positives = 40/89 (44%), Gaps = 4/89 (4%)
Frame = +2
Query: 35 LTGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEI----VKPNLQYHKQPGAS 90
L P+ +A+ + ++ P K TPP P L + V P+ + PG S
Sbjct: 170 LASPASASASDDNNNSRGVPKFKSTPP----------PSLPLSPPPVSPSSYFSIPPGLS 319
Query: 91 PSSKSRNSSFPSSPVSGSGSSSLLPSPTT 119
P+ F SPV SS++LPSPTT
Sbjct: 320 PAE------FLDSPVM-LNSSNILPSPTT 385
>TC79910 similar to GP|9293979|dbj|BAB01882.1 gene_id:MAL21.25~unknown
protein {Arabidopsis thaliana}, partial (67%)
Length = 741
Score = 34.7 bits (78), Expect = 0.023
Identities = 17/35 (48%), Positives = 23/35 (65%)
Frame = +3
Query: 82 QYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPS 116
Q QP +SPSS S +SSFPS P+ +++L PS
Sbjct: 126 QLKLQPSSSPSSLSFSSSFPSLPIISISTTTLSPS 230
>TC88770 homologue to GP|22202731|dbj|BAC07388. putative Rad6 {Oryza sativa
(japonica cultivar-group)}, partial (25%)
Length = 803
Score = 32.7 bits (73), Expect = 0.087
Identities = 28/106 (26%), Positives = 49/106 (45%), Gaps = 6/106 (5%)
Frame = +1
Query: 21 VHTNTGAFREVVQRLTGPSEGNATKEEQ-----TAKVAPITKRTPPKLHERRKCMKPKLE 75
+H F +VVQRLTGPS G ++ A++A I + +P + ++ + L
Sbjct: 121 LHIPVSDFMDVVQRLTGPSAGEEAPQQSGAVSPAARLASIERTSPTERERIQQTVDEDLT 300
Query: 76 IVKPNLQYHKQPG-ASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTP 120
+ ++ + PG SP+ ++ P P+S S + TTP
Sbjct: 301 WLLEGVEMGQFPGILSPAP----ATLP--PISQGFFSPMNEPQTTP 420
>TC88769 homologue to GP|19881561|gb|AAM00962.1 hypothetical protein {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (7%)
Length = 912
Score = 32.7 bits (73), Expect = 0.087
Identities = 28/106 (26%), Positives = 49/106 (45%), Gaps = 6/106 (5%)
Frame = +3
Query: 21 VHTNTGAFREVVQRLTGPSEGNATKEEQ-----TAKVAPITKRTPPKLHERRKCMKPKLE 75
+H F +VVQRLTGPS G ++ A++A I + +P + ++ + L
Sbjct: 276 LHIPVSDFMDVVQRLTGPSAGEEAPQQSGAVSPAARLASIERTSPTERERIQQTVDEDLT 455
Query: 76 IVKPNLQYHKQPG-ASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTP 120
+ ++ + PG SP+ ++ P P+S S + TTP
Sbjct: 456 WLLEGVEMGQFPGILSPAP----ATLP--PISQGFFSPMNEPQTTP 575
>TC92816 weakly similar to GP|18076679|emb|CAC84774. P70 protein {Nicotiana
tabacum}, partial (21%)
Length = 628
Score = 32.7 bits (73), Expect = 0.087
Identities = 27/97 (27%), Positives = 44/97 (44%), Gaps = 13/97 (13%)
Frame = +3
Query: 72 PKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSP--TTPSTLFSRLTI 129
PK P + K P A+P R + +P SGS S+S P+P TP + R+
Sbjct: 171 PKARSSAPEMSQRKSPAATP----RTARQLKTPNSGSNSASSSPNPIRKTPKDMSPRVNE 338
Query: 130 ME------DEKKQGSVINEI-----NIEEEEKAIKER 155
EKK+ S + E+ ++E+ K+ K++
Sbjct: 339 RRLSHSPISEKKRPSKVQELESQIAKLQEDLKSAKDQ 449
>BQ752445 similar to GP|18376387|em putative protein {Neurospora crassa},
partial (40%)
Length = 693
Score = 31.6 bits (70), Expect = 0.19
Identities = 16/44 (36%), Positives = 25/44 (56%)
Frame = +1
Query: 78 KPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
+ +L+ + PG +PSS S P+S + SS+ SPTTP+
Sbjct: 343 RTSLRPSRAPGRAPSSSPALRSRPASATGTTTSSTATTSPTTPA 474
>TC82331 weakly similar to EGAD|120604|128852 hypothetical protein M01F1.5
{Caenorhabditis elegans}, partial (10%)
Length = 848
Score = 31.6 bits (70), Expect = 0.19
Identities = 20/39 (51%), Positives = 25/39 (63%)
Frame = +1
Query: 87 PGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFS 125
P +SPS+ R S+ PSSP + S S S SPT+ STL S
Sbjct: 619 PCSSPSALPRRSAVPSSPHTSS*SLSAFSSPTS-STLES 732
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 31.6 bits (70), Expect = 0.19
Identities = 19/39 (48%), Positives = 24/39 (60%)
Frame = -1
Query: 87 PGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFS 125
P +SPSS +SS PSS S + SSS S ++ S LFS
Sbjct: 421 PSSSPSSPE*SSSSPSSSSSCTSSSSSSSSSSSVSLLFS 305
Score = 27.3 bits (59), Expect = 3.7
Identities = 17/34 (50%), Positives = 23/34 (67%), Gaps = 2/34 (5%)
Frame = -1
Query: 89 ASPSSKSRN-SSFPSSPV-SGSGSSSLLPSPTTP 120
+SPSS S + SS P +P+ SGS S S SP++P
Sbjct: 499 SSPSSSSSSPSSLPPAPLASGSSSGSPSSSPSSP 398
Score = 27.3 bits (59), Expect = 3.7
Identities = 19/38 (50%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Frame = -1
Query: 89 ASPSSKSRNSSFPSSPVSGS-GSSSLLPSPTTPSTLFS 125
+S SS S +SS PSS S S SSS PS P+ L S
Sbjct: 553 SSSSSSSSSSSSPSSSSSSSPSSSSSSPSSLPPAPLAS 440
>TC88983 similar to SP|P93231|VP41_LYCES Vacuolar assembly protein VPS41
homolog. [Tomato] {Lycopersicon esculentum}, partial
(17%)
Length = 668
Score = 31.6 bits (70), Expect = 0.19
Identities = 18/33 (54%), Positives = 20/33 (60%)
Frame = -3
Query: 88 GASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTP 120
G+S SS S SS SS S S SSS SP+TP
Sbjct: 231 GSSSSSSSSTSSSSSSSSSSSSSSSRSSSPSTP 133
Score = 27.7 bits (60), Expect = 2.8
Identities = 17/34 (50%), Positives = 20/34 (58%)
Frame = -3
Query: 92 SSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFS 125
SS S +SS SS S S SSS ++PST FS
Sbjct: 228 SSSSSSSSTSSSSSSSSSSSSSSSRSSSPSTPFS 127
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 31.2 bits (69), Expect = 0.25
Identities = 29/93 (31%), Positives = 37/93 (39%), Gaps = 15/93 (16%)
Frame = -2
Query: 91 PSSKSRNSSFPS----SPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVIN----- 141
P S NSS PS SP S S SSS LPS + S S L ++ G +N
Sbjct: 511 PPSSDDNSSSPSPNAPSPSSSSSSSSSLPSSPSASCCLS*LWVLLSPLVVGMSVNINA*S 332
Query: 142 ------EINIEEEEKAIKERRFYLHPSPRSKAS 168
+ I E + Y++ RS AS
Sbjct: 331 SGAKSVALTIAEANMVTRASAAYVNAPKRSVAS 233
Score = 26.2 bits (56), Expect = 8.2
Identities = 18/42 (42%), Positives = 23/42 (53%)
Frame = -2
Query: 89 ASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIM 130
+S SS S + S PSS S S SS P+P PS F L ++
Sbjct: 652 SSSSSSSSSWSSPSSSSSSSPFSSAPPAP--PSGAFFELLLL 533
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 31.2 bits (69), Expect = 0.25
Identities = 18/33 (54%), Positives = 21/33 (63%)
Frame = -2
Query: 89 ASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
+SPSS S +SS SS S S SSS+ PS PS
Sbjct: 429 SSPSSSSSSSSSSSSSSSSSSSSSIEPSFGAPS 331
Score = 26.2 bits (56), Expect = 8.2
Identities = 15/35 (42%), Positives = 21/35 (59%)
Frame = -2
Query: 89 ASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTL 123
+S S S +SS PSS S S SSS S ++ S++
Sbjct: 459 SSSSXSSSSSSSPSSSSSSSSSSSSSSSSSSSSSI 355
>TC80309 similar to GP|20804963|dbj|BAB92640. hypothetical protein~similar
to Arabidopsis thaliana chromosome 3 At3g51270, partial
(3%)
Length = 828
Score = 30.8 bits (68), Expect = 0.33
Identities = 24/81 (29%), Positives = 33/81 (40%), Gaps = 5/81 (6%)
Frame = -2
Query: 61 PKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGS-----GSSSLLP 115
P H+ + +L + G SP S S S F S S S SSS LP
Sbjct: 326 PLFHDFHVPHPAPTNVKNQHLNHLLHSGFSPRSPSSLSEFVSLSTSPSINL*ISSSSFLP 147
Query: 116 SPTTPSTLFSRLTIMEDEKKQ 136
P S+L + ++ E+KQ
Sbjct: 146 KPLAASSLSRKPALLSIEEKQ 84
>TC88544 similar to PIR|D96838|D96838 unknown protein T21F11.5 [imported] -
Arabidopsis thaliana, partial (30%)
Length = 1668
Score = 30.8 bits (68), Expect = 0.33
Identities = 23/87 (26%), Positives = 42/87 (47%)
Frame = +2
Query: 85 KQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEIN 144
K+P S S + SSF P S + S L PS + S++ ++ ++ + + +V + +
Sbjct: 149 KRPSFSESLRQVRSSFKQQPTSNTTPSPLSPSKQSESSV-DIMSKIQSFRYKTTVPDSDD 325
Query: 145 IEEEEKAIKERRFYLHPSPRSKASGFN 171
+ ++K I + Y RSK S N
Sbjct: 326 LLTQKKTISFQDMYSRMHNRSKDSAAN 406
>BQ751144 weakly similar to PIR|PQ0479|PQ04 pistil extensin-like protein
(clone pMG14) - common tobacco (fragment), partial (11%)
Length = 632
Score = 30.4 bits (67), Expect = 0.43
Identities = 19/37 (51%), Positives = 22/37 (59%)
Frame = +1
Query: 87 PGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTL 123
P +SPSS +R SS SP S S S P P+TPS L
Sbjct: 271 PASSPSSPTRPSSL-FSPPPPSPSRSRPPPPSTPSPL 378
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,899,217
Number of Sequences: 36976
Number of extensions: 78460
Number of successful extensions: 851
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 799
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 835
length of query: 190
length of database: 9,014,727
effective HSP length: 91
effective length of query: 99
effective length of database: 5,649,911
effective search space: 559341189
effective search space used: 559341189
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148755.1


