

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148654.4 + phase: 0
(246 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
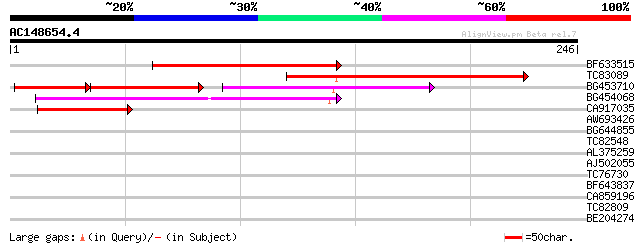
Score E
Sequences producing significant alignments: (bits) Value
BF633515 153 5e-38
TC83089 130 4e-31
BG453710 weakly similar to GP|21537057|gb| unknown {Arabidopsis ... 70 1e-30
BG454068 weakly similar to GP|22202804|dbj hypothetical protein~... 114 3e-26
CA917035 similar to GP|7303305|gb|A CG6061-PA {Drosophila melano... 45 3e-05
AW693426 homologue to GP|23307580|dbj similar to serine/threonin... 37 0.005
BG644855 weakly similar to GP|5777621|emb CAA30379.1 protein {Or... 28 4.2
TC82548 similar to GP|18253037|gb|AAL62445.1 unknown protein {Ar... 27 5.5
AL375259 weakly similar to GP|8843752|dbj| gene_id:MXK3.11~unkno... 27 7.2
AJ502055 similar to GP|4204304|gb|A lcl|prt_seq No definition li... 27 7.2
TC76730 similar to GP|20521241|dbj|BAB91757. putative 60S riboso... 27 7.2
BF643837 weakly similar to GP|22535591|db putative DEIH-box RNA/... 27 9.4
CA859196 weakly similar to PIR|C34715|R6BY 60s acidic ribosomal ... 27 9.4
TC82809 27 9.4
BE204274 weakly similar to GP|10177848|db gene_id:MCD7.26~pir||T... 27 9.4
>BF633515
Length = 249
Score = 153 bits (387), Expect = 5e-38
Identities = 74/82 (90%), Positives = 75/82 (91%)
Frame = +2
Query: 63 LDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSL 122
LDDVACLTHLPIEGRMLSH KKM KH+GA LLMRHLGVSQQEAEKICGQEYGGYISYP L
Sbjct: 2 LDDVACLTHLPIEGRMLSHGKKMPKHEGAALLMRHLGVSQQEAEKICGQEYGGYISYPRL 181
Query: 123 MDFYTTYLGRANLLAGTEDPEE 144
DFYTTYLGRANLLA TEDP E
Sbjct: 182 RDFYTTYLGRANLLASTEDPVE 247
>TC83089
Length = 887
Score = 130 bits (328), Expect = 4e-31
Identities = 67/108 (62%), Positives = 81/108 (74%), Gaps = 3/108 (2%)
Frame = +3
Query: 121 SLMDFYTTYLGRANLLAGTE---DPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELV 177
+L ++Y YL A L+ + DP+EL ++R CV+ YLLYLV CL FGD+SNKRIELV
Sbjct: 261 ALREYYEGYLDSATRLSDPQNLGDPQELEKIRTTCVKWYLLYLVGCLLFGDKSNKRIELV 440
Query: 178 YLTTMEEGYAGMRNYSWGGMTLAYLYGELAEACRPGDRALGGSVTLLT 225
YLTTME+GYAGMRN+SWGGMTLAYLY L+E PG AL SVTL+T
Sbjct: 441 YLTTMEDGYAGMRNHSWGGMTLAYLYHCLSEVSLPGGGALERSVTLVT 584
>BG453710 weakly similar to GP|21537057|gb| unknown {Arabidopsis thaliana},
partial (6%)
Length = 651
Score = 69.7 bits (169), Expect(3) = 1e-30
Identities = 27/49 (55%), Positives = 39/49 (79%)
Frame = -1
Query: 36 ALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKK 84
+++ +CE WH ETS+FH+P+GEMT+TLDDV L H+PI G ML H+++
Sbjct: 468 SMIRAMCERWHTETSSFHLPMGEMTITLDDVYNLLHIPIHGCMLDHDEQ 322
Score = 62.8 bits (151), Expect(3) = 1e-30
Identities = 37/95 (38%), Positives = 53/95 (54%), Gaps = 3/95 (3%)
Frame = -3
Query: 93 LLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRANLLAGT---EDPEELVRVR 149
L+ R LG+S A E G+ISYP+L Y +L A L E+ +E R R
Sbjct: 298 LMTRLLGMSDANAMAEIRTESAGHISYPTLKRVYEDHLIEARRLEDPLTREELQERARRR 119
Query: 150 PYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEE 184
+CVR LLYLV+C+ F ++N+ I+L+YL M +
Sbjct: 118 QWCVRSLLLYLVDCVLFTYKTNRHIDLIYLDCMAD 14
Score = 37.7 bits (86), Expect(3) = 1e-30
Identities = 16/33 (48%), Positives = 20/33 (60%)
Frame = -2
Query: 3 LRRPNGNENWFWDPLRESGLHDLVYLGYATVPH 35
L +P WF + + SGLHDL+Y GY TV H
Sbjct: 566 LGKPEACLRWFLELVEGSGLHDLIYTGYFTVTH 468
>BG454068 weakly similar to GP|22202804|dbj hypothetical protein~predicted by
GeneMark.hmm etc. {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 673
Score = 114 bits (285), Expect = 3e-26
Identities = 55/136 (40%), Positives = 81/136 (59%), Gaps = 3/136 (2%)
Frame = +3
Query: 12 WFWDPLRESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTH 71
WFW+ +R SGL+ L+ Y V H LL+ WH ETS+FH+P+GEMT+ LDD++CL H
Sbjct: 261 WFWNVIRASGLYPLLETNYGQVDHGLLIAFSLRWHSETSSFHLPVGEMTIALDDISCLLH 440
Query: 72 LPIEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLG 131
+P+ G +L HE + H+G L+ +LG+ +E + +I+Y +L+ YT+YL
Sbjct: 441 IPVGGNLLFHESLSI-HQGTEYLVNYLGLEFEEIAAETKRLKSAHITYDTLLSIYTSYLT 617
Query: 132 RANLLA---GTEDPEE 144
A A G ED E
Sbjct: 618 EAKSYANQPGEEDSME 665
>CA917035 similar to GP|7303305|gb|A CG6061-PA {Drosophila melanogaster},
partial (2%)
Length = 365
Score = 44.7 bits (104), Expect = 3e-05
Identities = 18/41 (43%), Positives = 26/41 (62%)
Frame = +2
Query: 13 FWDPLRESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFH 53
FW+ + SGL+ L+ Y V H+LL+ + WH ETS+FH
Sbjct: 242 FWNVIMASGLYLLLETNYGQVDHSLLIAFSDRWHSETSSFH 364
>AW693426 homologue to GP|23307580|dbj similar to serine/threonine
phosphatase PP7 {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 209
Score = 37.4 bits (85), Expect = 0.005
Identities = 22/69 (31%), Positives = 34/69 (48%)
Frame = +1
Query: 156 YLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLAYLYGELAEACRPGDR 215
+LL LV C F D+S + + YL + Y+WG LA+LY L A +
Sbjct: 1 FLLCLVGCTLFSDKSTFVVNVAYLEFFRD-LDSCGGYAWGVAALAHLYDNLRYASFHHMK 177
Query: 216 ALGGSVTLL 224
++ G +TL+
Sbjct: 178 SISGYLTLI 204
>BG644855 weakly similar to GP|5777621|emb CAA30379.1 protein {Oryza sativa},
partial (12%)
Length = 716
Score = 27.7 bits (60), Expect = 4.2
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 2/28 (7%)
Frame = -3
Query: 216 ALGGSVTLLTVRKLIFYFFVNFY--FFC 241
+LGGS+ L+ + +IF +NFY F+C
Sbjct: 174 SLGGSLPLICTQHVIFLLDINFYVRFYC 91
>TC82548 similar to GP|18253037|gb|AAL62445.1 unknown protein {Arabidopsis
thaliana}, partial (23%)
Length = 731
Score = 27.3 bits (59), Expect = 5.5
Identities = 10/29 (34%), Positives = 17/29 (58%)
Frame = -2
Query: 25 LVYLGYATVPHALLMTLCEMWHPETSTFH 53
+ ++ + H LL++LC+ HP S FH
Sbjct: 712 VTFISXLNLFHGLLLSLCQSPHPAFSDFH 626
>AL375259 weakly similar to GP|8843752|dbj| gene_id:MXK3.11~unknown protein
{Arabidopsis thaliana}, partial (22%)
Length = 466
Score = 26.9 bits (58), Expect = 7.2
Identities = 8/12 (66%), Positives = 11/12 (91%)
Frame = +2
Query: 231 FYFFVNFYFFCY 242
F+F+ +FYFFCY
Sbjct: 89 FFFYPSFYFFCY 124
>AJ502055 similar to GP|4204304|gb|A lcl|prt_seq No definition line found
{Arabidopsis thaliana}, partial (14%)
Length = 559
Score = 26.9 bits (58), Expect = 7.2
Identities = 20/65 (30%), Positives = 26/65 (39%)
Frame = +3
Query: 46 HPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMRHLGVSQQEA 105
HP T P + T CL+ +E RM + LK G+TLL R +E
Sbjct: 384 HPSTLWSLTPQSQTT-------CLSDSMLEHRMKILLSRSLKLTGSTLLARDSNAYPKEV 542
Query: 106 EKICG 110
CG
Sbjct: 543 SCTCG 557
>TC76730 similar to GP|20521241|dbj|BAB91757. putative 60S ribosomal protein
{Oryza sativa (japonica cultivar-group)}, partial (90%)
Length = 974
Score = 26.9 bits (58), Expect = 7.2
Identities = 14/42 (33%), Positives = 21/42 (49%), Gaps = 1/42 (2%)
Frame = -2
Query: 204 GELAEACRPGDRALGGSVTLLTVRKLIFYFFVNFY-FFCYIF 244
G L RPG + G + + K +YFF NF+ F ++F
Sbjct: 502 GNLPALLRPGPKRRGICLITASEAKKAWYFFANFFTSFLFLF 377
>BF643837 weakly similar to GP|22535591|db putative DEIH-box RNA/DNA helicase
{Oryza sativa (japonica cultivar-group)}, partial (3%)
Length = 469
Score = 26.6 bits (57), Expect = 9.4
Identities = 10/19 (52%), Positives = 11/19 (57%)
Frame = +1
Query: 150 PYCVRCYLLYLVECLWFGD 168
P+CV C YLV W GD
Sbjct: 70 PWCVACRFWYLVCIQWLGD 126
>CA859196 weakly similar to PIR|C34715|R6BY 60s acidic ribosomal protein
p1-alpha - fission yeast (Schizosaccharomyces pombe),
partial (88%)
Length = 512
Score = 26.6 bits (57), Expect = 9.4
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = -2
Query: 224 LTVRKLIFYFFVNFYFFCYIFR 245
+ +R F+F F+FFC+ FR
Sbjct: 373 IIIRFFFFFFSRFFFFFCFFFR 308
>TC82809
Length = 691
Score = 26.6 bits (57), Expect = 9.4
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = +1
Query: 232 YFFVNFYFFCYIFRA 246
YFF + YFFC +FR+
Sbjct: 271 YFFSSAYFFCVLFRS 315
>BE204274 weakly similar to GP|10177848|db gene_id:MCD7.26~pir||T02677~strong
similarity to unknown protein {Arabidopsis thaliana},
partial (22%)
Length = 392
Score = 26.6 bits (57), Expect = 9.4
Identities = 10/22 (45%), Positives = 15/22 (67%)
Frame = -1
Query: 225 TVRKLIFYFFVNFYFFCYIFRA 246
T++ F+F +NFY+ IFRA
Sbjct: 269 TLQCYCFFFHINFYYLVEIFRA 204
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,267,782
Number of Sequences: 36976
Number of extensions: 138145
Number of successful extensions: 1601
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 1486
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1566
length of query: 246
length of database: 9,014,727
effective HSP length: 94
effective length of query: 152
effective length of database: 5,538,983
effective search space: 841925416
effective search space used: 841925416
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148654.4


