

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148654.3 - phase: 0 /pseudo
(719 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
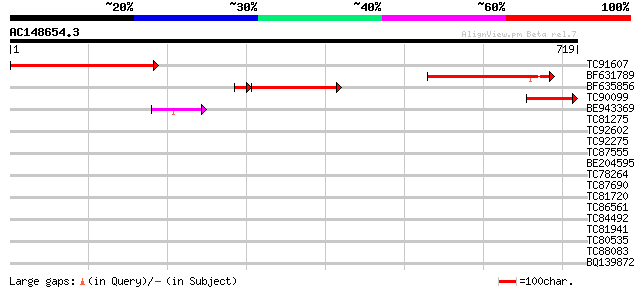
Score E
Sequences producing significant alignments: (bits) Value
TC91607 similar to GP|13548330|emb|CAC35877. putative protein {A... 358 3e-99
BF631789 similar to GP|13548330|em putative protein {Arabidopsis... 288 7e-78
BF635856 similar to GP|13548330|em putative protein {Arabidopsis... 232 6e-69
TC90099 similar to GP|13548330|emb|CAC35877. putative protein {A... 120 1e-27
BE943369 similar to GP|3859597|gb|A T15B16.3 gene product {Arabi... 47 3e-05
TC81275 similar to PIR|E84833|E84833 hypothetical protein At2g40... 32 0.85
TC92602 similar to SP|P57997|IF2C_PHAVU Translation initiation f... 32 1.1
TC92275 32 1.1
TC87555 weakly similar to GP|22775656|dbj|BAC15510. contains EST... 31 1.5
BE204595 similar to PIR|T48577|T485 hypothetical protein T31B5.1... 31 1.9
TC78264 homologue to SP|O48902|MDHP_MEDSA Malate dehydrogenase [... 30 3.2
TC87690 similar to GP|15028227|gb|AAK76610.1 putative oligopepti... 30 3.2
TC81720 similar to GP|8101699|gb|AAF72619.1| putative cellulose ... 30 4.2
TC86561 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-... 29 5.5
TC84492 similar to GP|23498276|emb|CAD49247. erythrocyte membran... 29 5.5
TC81941 similar to GP|15420162|gb|AAK97303.1 F-box containing pr... 29 7.2
TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9... 29 7.2
TC88083 similar to GP|4587373|dbj|BAA76734.1 ethylene responsive... 29 7.2
BQ139872 similar to GP|18376089|emb related to SOK1 protein {Neu... 29 7.2
>TC91607 similar to GP|13548330|emb|CAC35877. putative protein {Arabidopsis
thaliana}, partial (9%)
Length = 636
Score = 358 bits (920), Expect = 3e-99
Identities = 177/189 (93%), Positives = 179/189 (94%), Gaps = 1/189 (0%)
Frame = +2
Query: 1 MPTCHETSSTTTQLRLHTLTLTSMITACRASSSRANPLSLRSSLHSKPSFLSLSLFFPRH 60
MPTCHETSSTTTQLRLHTLTLTSMITACRASSSRANPLSLRSSLHSKPSFLSLSLFFPRH
Sbjct: 68 MPTCHETSSTTTQLRLHTLTLTSMITACRASSSRANPLSLRSSLHSKPSFLSLSLFFPRH 247
Query: 61 FHKSIALSSTTQCKPRSHLGGNLNNGLEEDGDREVHCELQVVSWRERRVKAEISINADIN 120
FHKSIALSSTTQCKPRSHLGGNLNNGLEEDGDREVHCELQVVSWRERRVKAEISINADIN
Sbjct: 248 FHKSIALSSTTQCKPRSHLGGNLNNGLEEDGDREVHCELQVVSWRERRVKAEISINADIN 427
Query: 121 SVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYW-HIEARVVLDLQEL 179
SVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQR + MY H + VLDLQEL
Sbjct: 428 SVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQR*ISKGMYLAH*SSVFVLDLQEL 607
Query: 180 LNSEWDREL 188
LNSEWDREL
Sbjct: 608 LNSEWDREL 634
Score = 36.6 bits (83), Expect = 0.035
Identities = 26/78 (33%), Positives = 40/78 (50%), Gaps = 2/78 (2%)
Frame = +2
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAIS-KILSRDNNKVRILQEGCKGLLYM 415
R V A I++ A + VWN ++ YE L + +PNL S +I ++ + Q +Y+
Sbjct: 386 RRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQR*ISKGMYL 565
Query: 416 VLHARV-VLDLCEQLEQE 432
+ V VLDL E L E
Sbjct: 566 AH*SSVFVLDLQELLNSE 619
>BF631789 similar to GP|13548330|em putative protein {Arabidopsis thaliana},
partial (17%)
Length = 549
Score = 288 bits (736), Expect = 7e-78
Identities = 151/183 (82%), Positives = 155/183 (84%), Gaps = 22/183 (12%)
Frame = +2
Query: 531 NSSSADDISDCNVQSSSNQRSRVPGLQRDIEVLKSELLKFVAEYGQEGFMPMRKQLRLHG 590
NSSSADDISDCNVQSSSNQRSRVPGLQRDIEVLKSELLKFVAEYGQEGFMPMRKQLRLHG
Sbjct: 2 NSSSADDISDCNVQSSSNQRSRVPGLQRDIEVLKSELLKFVAEYGQEGFMPMRKQLRLHG 181
Query: 591 RVDIEKAITRMGGFRKIATIMNLSLAYKYRKPKGYWDNLENLQDEISRFQRCWGMDPSFM 650
RVDIEKAITRMGGFRKIATIMNLSLAYKYRKPKGYWDNLENLQDEISRFQRCWGMDPSFM
Sbjct: 182 RVDIEKAITRMGGFRKIATIMNLSLAYKYRKPKGYWDNLENLQDEISRFQRCWGMDPSFM 361
Query: 651 PSRKSFERV----------------------VSQGKEKKNQTGQQPWQG*EK*SQRISRC 688
PSRKSFERV +SQGKEK+++ PWQG K*SQRISRC
Sbjct: 362 PSRKSFERVGRFDIARALEKWGGLHEGFSPSISQGKEKRSRQ-DTPWQGIRK*SQRISRC 538
Query: 689 R** 691
R**
Sbjct: 539 R** 547
>BF635856 similar to GP|13548330|em putative protein {Arabidopsis thaliana},
partial (14%)
Length = 408
Score = 232 bits (591), Expect(2) = 6e-69
Identities = 114/114 (100%), Positives = 114/114 (100%)
Frame = +1
Query: 307 SISGSLPASSSELNSNWGIFGKVCSLDKPCVVDEVHLRRFDGLLENGGVHRCVVASITVK 366
SISGSLPASSSELNSNWGIFGKVCSLDKPCVVDEVHLRRFDGLLENGGVHRCVVASITVK
Sbjct: 67 SISGSLPASSSELNSNWGIFGKVCSLDKPCVVDEVHLRRFDGLLENGGVHRCVVASITVK 246
Query: 367 APVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMVLHAR 420
APVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMVLHAR
Sbjct: 247 APVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMVLHAR 408
Score = 48.1 bits (113), Expect(2) = 6e-69
Identities = 21/21 (100%), Positives = 21/21 (100%)
Frame = +3
Query: 286 NGSLCETDKLAPGQDKEGLDT 306
NGSLCETDKLAPGQDKEGLDT
Sbjct: 3 NGSLCETDKLAPGQDKEGLDT 65
Score = 38.9 bits (89), Expect = 0.007
Identities = 21/65 (32%), Positives = 35/65 (53%)
Frame = +1
Query: 107 RRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYW 166
R V A I++ A + VWN ++ YE L + +PNL S +I ++ + Q G + +Y
Sbjct: 217 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAIS-KILSRDNNKVRILQEGCKGLLYM 393
Query: 167 HIEAR 171
+ AR
Sbjct: 394 VLHAR 408
>TC90099 similar to GP|13548330|emb|CAC35877. putative protein {Arabidopsis
thaliana}, partial (4%)
Length = 680
Score = 120 bits (302), Expect = 1e-27
Identities = 60/64 (93%), Positives = 61/64 (94%)
Frame = +1
Query: 656 FERVVSQGKEKKNQTGQQPWQG*EK*SQRISRCR**DQCSI*TLYFSRHTQVAYRTKRVG 715
F +SQGKEKKNQTGQQPWQG*EK*SQRISRCR**DQCSI*TLYFSRHTQVAYRTKRVG
Sbjct: 7 FSPSISQGKEKKNQTGQQPWQG*EK*SQRISRCR**DQCSI*TLYFSRHTQVAYRTKRVG 186
Query: 716 HKLG 719
HKLG
Sbjct: 187 HKLG 198
>BE943369 similar to GP|3859597|gb|A T15B16.3 gene product {Arabidopsis
thaliana}, partial (24%)
Length = 475
Score = 47.0 bits (110), Expect = 3e-05
Identities = 26/80 (32%), Positives = 44/80 (54%), Gaps = 10/80 (12%)
Frame = -3
Query: 180 LNSEWDRELHFSMVDGDFKKFEGKWSV----------KSGTRSSSTNLSYEVNVIPRFNF 229
L S R++ F M++GDF+ F+GKWS+ S R +T LSY V+V P+
Sbjct: 470 LPSGTKRDIDFKMIEGDFQLFKGKWSILQTFSNGSCEDSPVREINTTLSYIVDVKPKMWL 291
Query: 230 PAIFLERIVRSDLPVNLRAL 249
P +E + +++ NL+++
Sbjct: 290 PVRLIEGRLCNEIKKNLKSV 231
Score = 35.8 bits (81), Expect = 0.059
Identities = 26/87 (29%), Positives = 44/87 (49%), Gaps = 11/87 (12%)
Frame = -3
Query: 430 EQEISFEQAEGDFDSFHGKWTFEQLGNH-----------HTLLKYSVDSKMRRDTFLSEA 478
+++I F+ EGDF F GKW+ Q ++ +T L Y VD K + +L
Sbjct: 455 KRDIDFKMIEGDFQLFKGKWSILQTFSNGSCEDSPVREINTTLSYIVDVKPK--MWLPVR 282
Query: 479 IMEEVIYEDLPSNLCAIRDYVENQKAS 505
++E + ++ NL ++R E QKA+
Sbjct: 281 LIEGRLCNEIKKNLKSVRG--EAQKAT 207
>TC81275 similar to PIR|E84833|E84833 hypothetical protein At2g40760
[imported] - Arabidopsis thaliana, partial (27%)
Length = 883
Score = 32.0 bits (71), Expect = 0.85
Identities = 32/90 (35%), Positives = 43/90 (47%), Gaps = 7/90 (7%)
Frame = +3
Query: 22 TSMITACRASSSRANPLSLRSSLHSK----PSFLSLSLFFPRHFHKSIALSSTTQCKPRS 77
T M+++ +SSS P S + +S PSF +LSLF H +L S T K S
Sbjct: 21 TRMLSSSSSSSSSTTPSSETRTAYSSSSPNPSFKNLSLFSNNH-----SLQSMTLSKCFS 185
Query: 78 HLGGNLN---NGLEEDGDREVHCELQVVSW 104
H G +GL E G+ E L VVS+
Sbjct: 186 HSGPTYAIPVSGLVESGEVE---SLVVVSF 266
>TC92602 similar to SP|P57997|IF2C_PHAVU Translation initiation factor IF-2
chloroplast precursor (PvIF2cp). [Kidney bean French
bean], partial (17%)
Length = 912
Score = 31.6 bits (70), Expect = 1.1
Identities = 17/58 (29%), Positives = 30/58 (51%)
Frame = -3
Query: 2 PTCHETSSTTTQLRLHTLTLTSMITACRASSSRANPLSLRSSLHSKPSFLSLSLFFPR 59
PT ++ + L ++++ A R S S + S +S HS+P+F SL++ F R
Sbjct: 538 PTPFTAAAIDAEASSRVLLFSTVLKASRMSPSSQSS*SSNASPHSRPAFTSLTISFTR 365
>TC92275
Length = 479
Score = 31.6 bits (70), Expect = 1.1
Identities = 18/51 (35%), Positives = 27/51 (52%)
Frame = +3
Query: 472 DTFLSEAIMEEVIYEDLPSNLCAIRDYVENQKASQFLEVCEQNTNSGQQII 522
DTFL A + E Y +N D N +QF+ VC+++ NSG ++I
Sbjct: 141 DTFLFVAKVTEQTYRKPDNNKKDENDD*FNHNHNQFIRVCKESWNSGVKVI 293
>TC87555 weakly similar to GP|22775656|dbj|BAC15510. contains EST
C27990(C53630)~similar to G-protein beta family {Oryza
sativa (japonica cultivar-group), partial (33%)
Length = 685
Score = 31.2 bits (69), Expect = 1.5
Identities = 22/68 (32%), Positives = 35/68 (51%)
Frame = -2
Query: 388 NLAISKILSRDNNKVRILQEGCKGLLYMVLHARVVLDLCEQLEQEISFEQAEGDFDSFHG 447
+L+ KIL R NK+ I Q+ K +M+L + L +++++ E E D DS+
Sbjct: 327 SLSKEKILKRHTNKLAINQK-IKRFGFMIL----IFGLEKRIDERGEAESGEIDRDSYG* 163
Query: 448 KWTFEQLG 455
W FE G
Sbjct: 162 AWNFEWFG 139
>BE204595 similar to PIR|T48577|T485 hypothetical protein T31B5.120 -
Arabidopsis thaliana, partial (5%)
Length = 626
Score = 30.8 bits (68), Expect = 1.9
Identities = 17/53 (32%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Frame = +2
Query: 489 PSNLCAIRDY-VENQKASQFLEVCEQNTNSGQQIILSGSGDDNNSSSADDISD 540
P NLC+ + + VEN + Q + C ++ + +I S DN S DDI +
Sbjct: 416 PVNLCSKKVFDVENPGSPQRISYCAGDSFNAASLINSNISSDNESQLTDDIQN 574
>TC78264 homologue to SP|O48902|MDHP_MEDSA Malate dehydrogenase [NADP]
chloroplast precursor (EC 1.1.1.82) (NADP-MDH).
[Alfalfa], partial (91%)
Length = 1783
Score = 30.0 bits (66), Expect = 3.2
Identities = 22/77 (28%), Positives = 36/77 (46%), Gaps = 7/77 (9%)
Frame = +1
Query: 3 TCHETSS-------TTTQLRLHTLTLTSMITACRASSSRANPLSLRSSLHSKPSFLSLSL 55
T ETSS TT ++ H L+++ ++ + ++ N ++ LHS LS
Sbjct: 85 TLLETSS*SLVFFITTLIIKYHKLSVS*ILRFFDMALTQLNNTYSKTQLHSSSQLSFLSR 264
Query: 56 FFPRHFHKSIALSSTTQ 72
PRH H ++A TQ
Sbjct: 265 TLPRHHHCTLAPLRRTQ 315
>TC87690 similar to GP|15028227|gb|AAK76610.1 putative oligopeptidase A
{Arabidopsis thaliana}, partial (30%)
Length = 1019
Score = 30.0 bits (66), Expect = 3.2
Identities = 16/47 (34%), Positives = 26/47 (55%)
Frame = +2
Query: 19 LTLTSMITACRASSSRANPLSLRSSLHSKPSFLSLSLFFPRHFHKSI 65
L +++M+ A R + + + S RS +H S S F P+H HKS+
Sbjct: 20 LLISNMLMATRLTLTFSLSRSSRSRIHPLLKRTSCSFFNPKHSHKSL 160
>TC81720 similar to GP|8101699|gb|AAF72619.1| putative cellulose synthase
catalytic subunit {Gossypium hirsutum}, partial (36%)
Length = 605
Score = 29.6 bits (65), Expect = 4.2
Identities = 24/86 (27%), Positives = 35/86 (39%), Gaps = 1/86 (1%)
Frame = -1
Query: 2 PTCHETSSTTTQLRLHTLTLTSMITACRASSSRANPLSLRSSLHSKPS-FLSLSLFFPRH 60
P C S L + L L +++ C+ S NP R L K + F+ S FP
Sbjct: 584 PLCLANLSFLALLSIFQLFL-AILHCCKILSPLPNPCQ*RFLLKKKYAIFVDCSPHFPHD 408
Query: 61 FHKSIALSSTTQCKPRSHLGGNLNNG 86
H+ L P +HL +L+ G
Sbjct: 407 *HRGSGLDHHDDS*PETHLQNHLHQG 330
>TC86561 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-like
{Arabidopsis thaliana}, partial (17%)
Length = 661
Score = 29.3 bits (64), Expect = 5.5
Identities = 22/76 (28%), Positives = 39/76 (50%), Gaps = 7/76 (9%)
Frame = +3
Query: 207 KSGTRSSSTNLSYEVNVI---PRF----NFPAIFLERIVRSDLPVNLRALAYRVERNLLG 259
++ T S S +L EV I P F + ++F E + P++ + AY E+N G
Sbjct: 144 QTATGSPSPSLKPEVGEIDTSPPFQSVKDAVSLFGEGAFSGERPIHKKPKAYSAEKNCNG 323
Query: 260 NQKLPQPEDDLHKTSL 275
+K +P+ ++ +TSL
Sbjct: 324 GRKHDKPDFNVMQTSL 371
>TC84492 similar to GP|23498276|emb|CAD49247. erythrocyte membrane protein 1
(PfEMP1) {Plasmodium falciparum 3D7}, partial (1%)
Length = 751
Score = 29.3 bits (64), Expect = 5.5
Identities = 20/63 (31%), Positives = 30/63 (46%), Gaps = 4/63 (6%)
Frame = +3
Query: 36 NPLSLRSSLHSKPSFLSLSLFFPRHFHKSIA----LSSTTQCKPRSHLGGNLNNGLEEDG 91
NPL LRS S + F +H HKS+A L ST +S + + N L+E+
Sbjct: 270 NPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDLLSTATEHRKSFVSHQIPNQLQEEE 449
Query: 92 DRE 94
+ +
Sbjct: 450 EED 458
>TC81941 similar to GP|15420162|gb|AAK97303.1 F-box containing protein ORE9
{Arabidopsis thaliana}, partial (35%)
Length = 1017
Score = 28.9 bits (63), Expect = 7.2
Identities = 17/48 (35%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Frame = -2
Query: 4 CHETSSTTTQLRLHTLTLTSMITAC--RASSSRANPLSLRSSLHSKPS 49
CH + T Q +HT T S C ++NPL+L S+H K S
Sbjct: 605 CHCFAQTNHQPPIHTHTFASSSCLCLLHIPHLQSNPLTLNRSVHPKCS 462
>TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9O13.24
{Arabidopsis thaliana}, partial (55%)
Length = 1727
Score = 28.9 bits (63), Expect = 7.2
Identities = 41/202 (20%), Positives = 83/202 (40%), Gaps = 21/202 (10%)
Frame = +2
Query: 421 VVLDLCEQLEQEISFEQAEGDFDSFHGKWTFEQLGNHHTLLKYSVDSKMRRDTFLSEAIM 480
++ DLC + + S E A+ D F H+ +++ + K D + +
Sbjct: 296 ILFDLCGKSK---SVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCKSMTD---ARRVF 457
Query: 481 EEVIYEDLPSNLCAIRDYVENQKASQFLEVCEQNTNSGQQI-------ILSGSGDDNNSS 533
+ + ++ S IR Y + + L++ EQ G +I +LS G
Sbjct: 458 DHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLSACG------ 619
Query: 534 SADDISDCNV--QSSSNQRSRVPGLQRDIEVL----KSELLKFVAEYGQE--------GF 579
SA+ + D + +S ++ PG++ + +L +S LK E+ ++ F
Sbjct: 620 SAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLPFEPTVTVF 799
Query: 580 MPMRKQLRLHGRVDIEKAITRM 601
++ R+HG VD+E + +
Sbjct: 800 ETLKNYARIHGDVDLEDHVEEL 865
>TC88083 similar to GP|4587373|dbj|BAA76734.1 ethylene responsive element
binding factor {Nicotiana tabacum}, partial (46%)
Length = 1214
Score = 28.9 bits (63), Expect = 7.2
Identities = 15/32 (46%), Positives = 19/32 (58%)
Frame = +1
Query: 526 SGDDNNSSSADDISDCNVQSSSNQRSRVPGLQ 557
SG D++SS DD DC + SSS R P +Q
Sbjct: 646 SGCDSSSSVVDDDEDCVILSSSAASVRKPPVQ 741
>BQ139872 similar to GP|18376089|emb related to SOK1 protein {Neurospora
crassa}, partial (6%)
Length = 658
Score = 28.9 bits (63), Expect = 7.2
Identities = 20/51 (39%), Positives = 24/51 (46%), Gaps = 4/51 (7%)
Frame = +1
Query: 3 TCHETSSTTTQLRLHTLTLTSMITACRASSSR----ANPLSLRSSLHSKPS 49
T + S T + T T T + A S+SR L LRSSLHS PS
Sbjct: 241 TSNHYSLRTQSISKRTTTWTGWLPAVPESTSRLRRSGTALRLRSSLHSAPS 393
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,131,833
Number of Sequences: 36976
Number of extensions: 346562
Number of successful extensions: 1848
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1802
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1843
length of query: 719
length of database: 9,014,727
effective HSP length: 103
effective length of query: 616
effective length of database: 5,206,199
effective search space: 3207018584
effective search space used: 3207018584
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148654.3


