

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148653.1 - phase: 0
(693 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
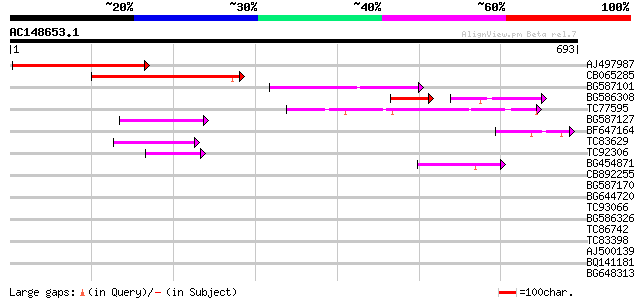
Score E
Sequences producing significant alignments: (bits) Value
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 335 3e-92
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 315 5e-86
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 108 9e-24
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 60 4e-18
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 73 3e-13
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 70 2e-12
BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis t... 50 4e-06
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 48 1e-05
TC92306 47 3e-05
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 44 3e-04
CB892255 40 0.002
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 40 0.002
BG644720 38 0.011
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 38 0.015
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 37 0.019
TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG00... 37 0.019
TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.... 35 0.13
AJ500139 similar to GP|10129650|emb histidine kinase-like protei... 32 1.1
BQ141181 similar to GP|8777430|dbj| dbj|BAA91947.1~gene_id:MHM17... 29 5.3
BG648313 similar to GP|9279604|dbj DNA-binding protein GT-1 {Ara... 29 5.3
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 335 bits (860), Expect = 3e-92
Identities = 154/167 (92%), Positives = 164/167 (97%)
Frame = -2
Query: 4 KTCCALAWAAKRLRHYMISHTTWLISKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 63
KTCCALAWAAKRLRHYMI+HTTWL+SKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 64 QKAIKGSILADHLAHQPIEDYQPIKFDFPDEEIMYLKMKDCDEPLLGEGPDPESRWGLIF 123
QKAIKGSILADHLAHQP+EDY+PIKFDFPDEEIMYLKMKDCDEPL GEGPDP+S WGLIF
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 124 DGAVNVYGSGIGAIIVTPEGAHIPFTARLTFDCTNNIAEYEACIMGI 170
DGAVNVYG+GIGA+++TP+GAHIPFTARL FDCTNNIAEYEACIMGI
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 315 bits (806), Expect = 5e-86
Identities = 145/196 (73%), Positives = 170/196 (85%), Gaps = 8/196 (4%)
Frame = -2
Query: 100 KMKDCDEPLLGEGPDPESRWGLIFDGAVNVYGSGIGAIIVTPEGAHIPFTARLTFDCTNN 159
K KDC+EPL+GEGPDP S+WGL+FDGAVN YG GIGA+IV+P+G +IPFTAR+ F+CTNN
Sbjct: 591 KSKDCEEPLIGEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNN 412
Query: 160 IAEYEACIMGIEEAIDLRIKNLDIYGDSALVINQITGEWETRHPGLVPYKDYARRLLTFF 219
+AEYEACI GIEEAID+RIK+LDIYGDSALVINQI GEWET H L+PY+DYARRLLT+F
Sbjct: 411 MAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYF 232
Query: 220 NKVELHHIPRDENQMADALATLSSMYQVNRWNEVPLIHIRHLERPAHVFAI--------E 271
KVELHHIPRDENQMADALATLSSM++VN WN+VP+I ++ LERP+HVFAI E
Sbjct: 231 TKVELHHIPRDENQMADALATLSSMFRVNHWNDVPIIKVQRLERPSHVFAIGNVIDQTGE 52
Query: 272 EVVDDKPWFHDIKCFL 287
VVD KPW++DIK FL
Sbjct: 51 NVVDYKPWYYDIKQFL 4
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 108 bits (269), Expect = 9e-24
Identities = 59/188 (31%), Positives = 96/188 (50%)
Frame = +2
Query: 318 LYKRNFDMVLLRCVDKHEADLLMHEIHEGSFGTHSSGPTMAKKMLRAGYYWMTMEADCYK 377
LYK+ D + +RCV + E ++ H ++ H + K+ +AG++W TM D +
Sbjct: 41 LYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHS 220
Query: 378 HAKRCYKCQIYADRIHVPPTALNVLSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYF 437
+C CQ + N + F +WGID +G +N ++ILVA+DY
Sbjct: 221 FISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPFPSSYNN--KYILVAVDYV 394
Query: 438 TKWVEAASYANVTKQVVVKFIKNHIICRYGVPSRIITDNGTNLNNKMMKELCDDFKIEHH 497
+KWVEA + VVVK K+ I R+GVP +I+D G++ NK+ ++L + H
Sbjct: 395 SKWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHK 574
Query: 498 NSSPYRPQ 505
++ Y PQ
Sbjct: 575 VATAYHPQ 598
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 60.5 bits (145), Expect(2) = 4e-18
Identities = 41/122 (33%), Positives = 67/122 (54%), Gaps = 4/122 (3%)
Frame = -3
Query: 539 LHGYRTSIRTSTGATPFSLVYGMEAVLPIEVEIPSI---RVLMEAELSEAEWVQSRYDQL 595
L +RT+ R +T +TPFS+ + +EA+ P EV + S+ R+ EL+ ++ L
Sbjct: 462 LWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSRMPQNIELNN----DRLFNAL 295
Query: 596 NLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQPD-SRGKWTPNY 654
IEE+R AL Q YQ +++ ++K V + K GD+VL K+ + + GK N+
Sbjct: 294 ETIEERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFENTKELNAGKLGTNW 115
Query: 655 EG 656
EG
Sbjct: 114 EG 109
Score = 49.3 bits (116), Expect(2) = 4e-18
Identities = 21/52 (40%), Positives = 34/52 (65%)
Frame = -2
Query: 466 YGVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNI 517
+G+P I+TDNG++ + +E C+ ++I + +SP PQ NG EA+NK I
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKII 530
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 73.2 bits (178), Expect = 3e-13
Identities = 72/325 (22%), Positives = 134/325 (41%), Gaps = 13/325 (4%)
Frame = +2
Query: 339 LMHEIHEGSFGTHSSGPTMAKKMLRAGYYWMTMEADCYKHAKRCYKCQIYADRIHVPPTA 398
L+ E H+ + H G +++ ++W + + C C IH+ A
Sbjct: 179 LVQESHDSTAAGHP-GRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVC----GGIHIWRQA 343
Query: 399 LNVLSSPWPF-----SMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYANVTKQV 453
P P S +D I + P G +++ V +D +K V + +
Sbjct: 344 KRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDTMEAEA 523
Query: 454 VVKFIKNHIICRY---GVPSRIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAV 510
+ + C Y G+P I++D G+N + +E C + S+ Y PQ +G
Sbjct: 524 CAQ---RFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGT 694
Query: 511 EAANKNIKKIIQKMVVTYKD-WHEMLPFALHGYRTSIRTSTGATPFSLVYGMEAVLPIEV 569
E N+ I+ +++ V +D W ++LP R +S GATPF + +G V PI
Sbjct: 695 ERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYH-VDPIPT 871
Query: 570 EIPSIRVLMEAELSEAEWVQSRYDQLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPRE- 628
+ V+ E E + V+ D I+ + ++A Q+R + + +K+ P +
Sbjct: 872 VEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAA-------QQRSEASANKRRCPADR 1030
Query: 629 FKEGDLVLKKILTF---QPDSRGKW 650
++ GD V + + +P + W
Sbjct: 1031YQVGDKVWLNVSNYKSPRPSKKLDW 1105
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 70.5 bits (171), Expect = 2e-12
Identities = 41/109 (37%), Positives = 58/109 (52%)
Frame = +3
Query: 135 GAIIVTPEGAHIPFTARLTFDCTNNIAEYEACIMGIEEAIDLRIKNLDIYGDSALVINQI 194
G + +P + + RL F +NN YEA I G+ A L+I+N+ Y DS LV +Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 195 TGEWETRHPGLVPYKDYARRLLTFFNKVELHHIPRDENQMADALATLSS 243
+GE+E R + Y ++L + L IPR EN ADALA L+S
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALAS 329
>BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana},
partial (6%)
Length = 469
Score = 49.7 bits (117), Expect = 4e-06
Identities = 32/107 (29%), Positives = 57/107 (52%), Gaps = 10/107 (9%)
Frame = +1
Query: 594 QLNLIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVL---KKILTFQPDSRGKW 650
+LN +EE R+ A + ++Y++R K D+++ REF+EG+LVL ++ F R W
Sbjct: 103 ELNELEELRLDAYENAKIYKERTKKWHDRRIIRREFREGELVLLFNSRLKLFPGKLRSHW 282
Query: 651 TPNYEGPYVVKKAYSGGAMTLQ-------TMDGEELQRPVNTDAVKK 690
+ GP+ VK GA+ + T++G+ L+ + + V K
Sbjct: 283 S----GPFQVKNVMPSGAVEVWSESTGPFTVNGQRLKHYTSGENVDK 411
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 47.8 bits (112), Expect = 1e-05
Identities = 31/106 (29%), Positives = 50/106 (46%)
Frame = +2
Query: 127 VNVYGSGIGAIIVTPEGAHIPFTARLTFDCTNNIAEYEACIMGIEEAIDLRIKNLDIYGD 186
+N +G GA+++ +G+ + + T AEY A ++G++ A K + GD
Sbjct: 584 INRGPAGAGALLLAEDGSLLYGFRQGLGHQTKESAEYRALLLGLKHASMKGFKYVTAKGD 763
Query: 187 SALVINQITGEWETRHPGLVPYKDYARRLLTFFNKVELHHIPRDEN 232
S LVINQI W+ + L A L F+ + HI R+ N
Sbjct: 764 SELVINQILDPWKIKDEHLKKLCAEALELSDNFHSFRIQHISRERN 901
>TC92306
Length = 521
Score = 46.6 bits (109), Expect = 3e-05
Identities = 24/73 (32%), Positives = 37/73 (49%)
Frame = +2
Query: 167 IMGIEEAIDLRIKNLDIYGDSALVINQITGEWETRHPGLVPYKDYARRLLTFFNKVELHH 226
I+G+ EA + +++ + GD V Q G W+ +P L + A L + F V + H
Sbjct: 2 ILGLNEARNQGYEHVHVRGDFQXVCKQFXGSWKVNNPNLRNLCNXAVELKSNFKSVSVEH 181
Query: 227 IPRDENQMADALA 239
+PR N ADA A
Sbjct: 182 VPRGXNXAADAQA 220
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 43.5 bits (101), Expect = 3e-04
Identities = 32/117 (27%), Positives = 55/117 (46%), Gaps = 9/117 (7%)
Frame = +2
Query: 499 SSPYRPQMNGAVEAANKNIKKIIQKMVVTYK-DWHEMLPFALHGYRTSIRTSTGATPFSL 557
SS Y P +G EA NK + ++ ++ T W + P+A + Y TS S TPF
Sbjct: 83 SSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEYWYNTSYNISAAMTPFKA 262
Query: 558 VYGMEAVLPIE--------VEIPSIRVLMEAELSEAEWVQSRYDQLNLIEEKRMSAL 606
+YG + + I ++ S E LS+ + + +R ++L I+ R +A+
Sbjct: 263 LYGRDLSMLIRSKGSSKDTADLQSQLAQREELLSQLQSISTRLNKL*SIKLIRNAAI 433
Score = 30.8 bits (68), Expect = 1.8
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Frame = +1
Query: 600 EKRMSALCHGQLY--QKRMKHAFDKKVHPREFKEGDLVLKKILTFQPDS 646
E+R++ LY Q+ MKH DKK EF+ G+ VL K+ +Q S
Sbjct: 346 ERRIAVTATIHLYKAQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSS 492
>CB892255
Length = 853
Score = 40.4 bits (93), Expect = 0.002
Identities = 19/34 (55%), Positives = 23/34 (66%)
Frame = +1
Query: 622 KKVHPREFKEGDLVLKKILTFQPDSRGKWTPNYE 655
KK PRE +E VLK +L P+SRGKW PNY+
Sbjct: 178 KKARPRECQESGFVLKNML---PNSRGKWMPNYK 270
Score = 38.9 bits (89), Expect = 0.007
Identities = 34/98 (34%), Positives = 42/98 (42%), Gaps = 1/98 (1%)
Frame = +2
Query: 597 LIEEKRMSALCHGQLYQKRMKHAFDKKVHPREFKEGDLVLKKILTFQPDSRGKWTPNYEG 656
LIE K + A CHGQ YQ+ +++ P K+ L LK
Sbjct: 104 LIELKSVIAGCHGQSYQEDKASIRQRRLVPVNVKKVALCLKICYPIL------------- 244
Query: 657 PYVVKKAYSGGAMTLQ-TMDGEELQRPVNTDAVKKYFV 693
GA Q T +G +L PVNTDAVKKYFV
Sbjct: 245 ----------GANGCQITKNGGKLVCPVNTDAVKKYFV 328
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 40.4 bits (93), Expect = 0.002
Identities = 26/105 (24%), Positives = 48/105 (44%), Gaps = 4/105 (3%)
Frame = -3
Query: 425 NGHRFILVAIDYFTKWVEAASYANVTKQVVVKFIKN---HIICRYGVPSRII-TDNGTNL 480
+ H++ + ID +K+ + K V+ KN ++ Y +I+ +DNG
Sbjct: 383 DNHKYFVTFIDEKSKYTWLTLIPS--KDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEY 210
Query: 481 NNKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNIKKIIQKMV 525
+ K D I H S PY PQ NG + NK++ ++ + ++
Sbjct: 209 TSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLM 75
>BG644720
Length = 678
Score = 38.1 bits (87), Expect = 0.011
Identities = 37/137 (27%), Positives = 58/137 (42%), Gaps = 1/137 (0%)
Frame = -3
Query: 292 YPLGASNKDKKT-LRRLSGNFFLNGDILYKRNFDMVLLRCVDKHEADLLMHEIHEGSFGT 350
Y + N + T +RR + + + LY F+ VLL C+ + E H G+
Sbjct: 433 YGIFLENPRRSTYIRRHTPHLH*YNETLYI*LFEGVLL*CLGEEETIQAFQ*AHSRVCGS 254
Query: 351 HSSGPTMAKKMLRAGYYWMTMEADCYKHAKRCYKCQIYADRIHVPPTALNVLSSPWPFSM 410
H S + + R GYYW T K + A+ I PP L+ + F
Sbjct: 253 HKS--KLYFHIKRMGYYWNT---------*IMLKGALLANFIQQPPEVLHPTITS*LFES 107
Query: 411 WGIDMIGRIEPKASNGH 427
W +++ G + PK+S GH
Sbjct: 106 WELNVFG-LFPKSSGGH 59
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 37.7 bits (86), Expect = 0.015
Identities = 28/101 (27%), Positives = 44/101 (42%), Gaps = 3/101 (2%)
Frame = +1
Query: 414 DMIGRIEPKASNGHRFILVAIDYFTK--WVEAASYANVTKQVVVKFIKNHIICRYGVP-S 470
D+ G + + G R+++ ID F + WV Y N T K+ + + + G
Sbjct: 115 DLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKW-RILVETQTGKNVK 291
Query: 471 RIITDNGTNLNNKMMKELCDDFKIEHHNSSPYRPQMNGAVE 511
++ITDN + E C + I H + P PQ NG E
Sbjct: 292 KLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAE 414
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 37.4 bits (85), Expect = 0.019
Identities = 20/75 (26%), Positives = 39/75 (51%)
Frame = +2
Query: 8 ALAWAAKRLRHYMISHTTWLISKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRSQKAI 67
A+ +A K R Y+ + + +KYIF +P L R RW +++YD++
Sbjct: 182 AVVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPG- 358
Query: 68 KGSILADHLAHQPIE 82
K +++AD L+ + ++
Sbjct: 359 KANLVADALSRRRVD 403
>TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG005I10.16 -
Arabidopsis thaliana, partial (19%)
Length = 2073
Score = 37.4 bits (85), Expect = 0.019
Identities = 28/86 (32%), Positives = 47/86 (54%), Gaps = 7/86 (8%)
Frame = -1
Query: 161 AEYEACIMGIEEAIDLRIKNLDIYGDSALVIN---QITGEWETRHPGLVPYKDYAR--RL 215
AE+ AC++ IE+A++L + N+ + DS V+N +I G +P++ R
Sbjct: 396 AEFCACMIAIEKAMELGLNNICLETDSLKVVNAFHKIVG---------IPWQMRVRWHNC 244
Query: 216 LTFFNKVE--LHHIPRDENQMADALA 239
+ F + + HIPR+ N +ADALA
Sbjct: 243 IRFCHSIACVCVHIPREGNLVADALA 166
>TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.40 -
Arabidopsis thaliana, partial (6%)
Length = 766
Score = 34.7 bits (78), Expect = 0.13
Identities = 31/93 (33%), Positives = 46/93 (49%), Gaps = 4/93 (4%)
Frame = +2
Query: 155 DCTNNI--AEYEACIMGIEEAIDLRIKNLDIYGDSALVINQITGEWETRHPGLVPYKDYA 212
D +N+I AE A +MG++ A L I ++ Y DS +N I G P +V Y YA
Sbjct: 122 DHSNDILFAELHAILMGLQLAQTLNIVDVVCYSDSLHYVNLING------PSVV-YHAYA 280
Query: 213 RRLLTFFNKVELH--HIPRDENQMADALATLSS 243
+ + + L H R+ N+ AD LA L +
Sbjct: 281 TLIQDIKDLIRLSKLHTLREGNRCADFLAKLGA 379
>AJ500139 similar to GP|10129650|emb histidine kinase-like protein
{Arabidopsis thaliana}, partial (6%)
Length = 475
Score = 31.6 bits (70), Expect = 1.1
Identities = 25/80 (31%), Positives = 36/80 (44%)
Frame = -2
Query: 373 ADCYKHAKRCYKCQIYADRIHVPPTALNVLSSPWPFSMWGIDMIGRIEPKASNGHRFILV 432
AD +K +++C CQ+Y R P S I RI PK GHR
Sbjct: 369 ADSHKFSQQC--CQVYTSR--------QSWYKPLCCSRTSICQRRRISPK---GHRR--- 238
Query: 433 AIDYFTKWVEAASYANVTKQ 452
++YF+KW+E +TK+
Sbjct: 237 PVNYFSKWIEGR*TYTITKK 178
>BQ141181 similar to GP|8777430|dbj| dbj|BAA91947.1~gene_id:MHM17.1~similar
to unknown protein {Arabidopsis thaliana}, partial (13%)
Length = 585
Score = 29.3 bits (64), Expect = 5.3
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 7/57 (12%)
Frame = -2
Query: 468 VPSRIITDNGTNLN-------NKMMKELCDDFKIEHHNSSPYRPQMNGAVEAANKNI 517
+P + +TDN LN NK+ + L N S +PQ+ ++EAANKN+
Sbjct: 410 LPDK*VTDNPKRLNFPLPFNQNKLSQTL---------NPSILKPQLTESLEAANKNL 267
>BG648313 similar to GP|9279604|dbj DNA-binding protein GT-1 {Arabidopsis
thaliana}, partial (53%)
Length = 815
Score = 29.3 bits (64), Expect = 5.3
Identities = 20/43 (46%), Positives = 24/43 (55%), Gaps = 3/43 (6%)
Frame = -3
Query: 115 PESRWGLIFDG---AVNVYGSGIGAIIVTPEGAHIPFTARLTF 154
P+S +FDG A+N Y S IVTP + P TARLTF
Sbjct: 798 PKSGPNCLFDGFRSAINTYSS----CIVTPFD*YNPSTARLTF 682
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,704,411
Number of Sequences: 36976
Number of extensions: 327653
Number of successful extensions: 1358
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1343
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1353
length of query: 693
length of database: 9,014,727
effective HSP length: 103
effective length of query: 590
effective length of database: 5,206,199
effective search space: 3071657410
effective search space used: 3071657410
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148653.1


