

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.4 + phase: 0 /pseudo
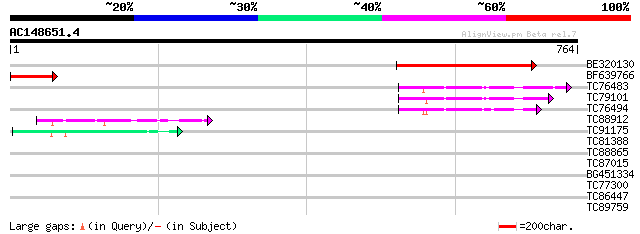
(764 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE320130 similar to GP|22128583|gb| molybdenum cofactor sulfuras... 356 2e-98
BF639766 similar to GP|22128583|gb| molybdenum cofactor sulfuras... 128 9e-30
TC76483 weakly similar to GP|9755390|gb|AAF98197.1| F17F8.22 {Ar... 75 9e-14
TC79101 weakly similar to GP|9755390|gb|AAF98197.1| F17F8.22 {Ar... 74 2e-13
TC76494 weakly similar to GP|9755390|gb|AAF98197.1| F17F8.22 {Ar... 61 1e-09
TC88912 similar to GP|16152176|gb|AAL14994.1 NIFS-like protein C... 44 3e-04
TC91175 weakly similar to PIR|T05129|T05129 hypothetical protein... 43 5e-04
TC81388 weakly similar to PIR|T05129|T05129 hypothetical protein... 37 0.028
TC88865 similar to PIR|H87729|H87729 protein Y23H5A.7 [imported]... 33 0.41
TC87015 33 0.53
BG451334 weakly similar to GP|9755390|gb| F17F8.22 {Arabidopsis ... 31 2.0
TC77300 similar to PIR|T48643|T48643 cinnamoyl CoA reductase-lik... 29 5.9
TC86447 weakly similar to GP|19911585|dbj|BAB86896. syringolide-... 29 5.9
TC89759 similar to GP|13430174|gb|AAK25754.1 putative chaperon P... 29 5.9
>BE320130 similar to GP|22128583|gb| molybdenum cofactor sulfurase
{Lycopersicon esculentum}, partial (6%)
Length = 588
Score = 356 bits (913), Expect = 2e-98
Identities = 179/190 (94%), Positives = 181/190 (95%), Gaps = 1/190 (0%)
Frame = +3
Query: 522 LATMGSLKHDREWILKSLSGEILTLKRVPEMGLISSFIDLSQGMLFVESPHCKERLQIRL 581
L+ GSLKHDREWILKSLSGEILTLKRVPEMGLISSFIDLSQGMLFVESPHCKERLQIRL
Sbjct: 3 LSNNGSLKHDREWILKSLSGEILTLKRVPEMGLISSFIDLSQGMLFVESPHCKERLQIRL 182
Query: 582 QLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDFVLDRTKDI 641
QLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDFVLDRTKDI
Sbjct: 183 QLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDFVLDRTKDI 362
Query: 642 VTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNRFRPN-LVV 700
VTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNRFRPN +
Sbjct: 363 VTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNRFRPNPCGI 542
Query: 701 SGGRPYDEDG 710
PYDEDG
Sbjct: 543 LEADPYDEDG 572
>BF639766 similar to GP|22128583|gb| molybdenum cofactor sulfurase
{Lycopersicon esculentum}, partial (7%)
Length = 298
Score = 128 bits (321), Expect = 9e-30
Identities = 63/64 (98%), Positives = 63/64 (98%)
Frame = +3
Query: 1 MAKEEFLKEFGEHYGYPNAARTIDQIRATEFNRLQDLVYLDHAGATLYSELQMESVFKDL 60
MAKE FLKEFGEHYGYPNAARTIDQIRATEFNRLQDLVYLDHAGATLYSELQMESVFKDL
Sbjct: 105 MAKE*FLKEFGEHYGYPNAARTIDQIRATEFNRLQDLVYLDHAGATLYSELQMESVFKDL 284
Query: 61 TTNV 64
TTNV
Sbjct: 285 TTNV 296
>TC76483 weakly similar to GP|9755390|gb|AAF98197.1| F17F8.22 {Arabidopsis
thaliana}, partial (61%)
Length = 1632
Score = 75.1 bits (183), Expect = 9e-14
Identities = 62/243 (25%), Positives = 106/243 (43%), Gaps = 10/243 (4%)
Frame = +1
Query: 524 TMGSLKHDREWILKSLSGEILTLKRVPEMGLIS----------SFIDLSQGMLFVESPHC 573
T + DR+W++ + G T + P++ L+ + + + +++P
Sbjct: 274 TPSGFRWDRQWMVVNSKGRACTQRVEPKLALVEVEFPPEAFDEHWEPTTDSFMVLKAPGM 453
Query: 574 KERLQIRLQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDF 633
E L++ L + DI + + E + WFS + P L+R++ +S
Sbjct: 454 -EPLKVCLNKQY--EVADDITVWEWTGSAWDEGAEASQWFSDYLGNPTKLVRFNTASEVR 624
Query: 634 VLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNR 693
+D D V + F++ FL+ S+ES+ LN+ L + M NR
Sbjct: 625 KVD--PDYV---EGQYQTFFSDGYPFLIASQESLDALNELLEEPILM-----------NR 756
Query: 694 FRPNLVVSGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQVINLTLNAGQVQKSKEPLATLA 753
FRPN++V G PY ED W D +I + F+ C RC++ T+N + EP TL
Sbjct: 757 FRPNILVEGCEPYSEDLWRDFKISSFSFQGAKLCYRCKI--PTINQETAKVGPEPNETLM 930
Query: 754 SYR 756
YR
Sbjct: 931 KYR 939
>TC79101 weakly similar to GP|9755390|gb|AAF98197.1| F17F8.22 {Arabidopsis
thaliana}, partial (76%)
Length = 1119
Score = 74.3 bits (181), Expect = 2e-13
Identities = 59/219 (26%), Positives = 96/219 (42%), Gaps = 10/219 (4%)
Frame = +2
Query: 524 TMGSLKHDREWILKSLSGEILTLKRVPEMGLISSFID----------LSQGMLFVESPHC 573
T L+ DR+W++ + G T + P++ L+ + + + +++P
Sbjct: 119 TPSGLRWDRQWVVVNSKGRACTQRVEPKLALVEVELPPEAFDEHWEPTTDSFMVLKAPGM 298
Query: 574 KERLQIRLQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDF 633
E L++ L + DI + + E WFS + P L+R++ +S
Sbjct: 299 -EPLKVFLNKQY--EVADDITVWEWTGSAWDEGAEAAQWFSDYLGNPTKLVRFNTASEVR 469
Query: 634 VLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNR 693
+D D V + F + FLLVS+ES+ LN L + M NR
Sbjct: 470 KVD--PDYV---EGQQQTFFTDGYPFLLVSQESLDALNAHLEEPIPM-----------NR 601
Query: 694 FRPNLVVSGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQV 732
FRPN++V G PY ED W DI+I F+ + C RC+V
Sbjct: 602 FRPNILVEGCEPYSEDLWRDIKISRFSFQGVKLCARCKV 718
>TC76494 weakly similar to GP|9755390|gb|AAF98197.1| F17F8.22 {Arabidopsis
thaliana}, partial (65%)
Length = 754
Score = 61.2 bits (147), Expect = 1e-09
Identities = 53/203 (26%), Positives = 90/203 (44%), Gaps = 10/203 (4%)
Frame = +3
Query: 524 TMGSLKHDREWILKSLSGEILTLKRVPEMGLIS------SFID----LSQGMLFVESPHC 573
T + DR+W++ + G T++ P LI +F+ + + +++P
Sbjct: 171 TPSGFRWDRQWMVVNSKGRACTIRVEPRFALIDVELPPEAFLQHWEPTTDSFMVLKAPGM 350
Query: 574 KERLQIRLQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDF 633
E L++ L + D+ + + E + WFS + P L+R++ +S
Sbjct: 351 -EPLKVCLNKQY--EVEDDVTVWEWTGSAWDEGAEASQWFSDYLGYPAKLVRFNTASEVR 521
Query: 634 VLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNR 693
+D D V + F++ FLLVS +S+ LN+ L + M NR
Sbjct: 522 KVD--PDYVKGQHQTF---FSDGYPFLLVSHDSLDSLNELLEEPIPM-----------NR 653
Query: 694 FRPNLVVSGGRPYDEDGWSDIRI 716
FRPN++V G PY ED W DI+I
Sbjct: 654 FRPNILVEGCGPYSEDLWRDIKI 722
>TC88912 similar to GP|16152176|gb|AAL14994.1 NIFS-like protein CpNifsp
precursor {Arabidopsis thaliana}, partial (52%)
Length = 991
Score = 43.5 bits (101), Expect = 3e-04
Identities = 58/247 (23%), Positives = 108/247 (43%), Gaps = 10/247 (4%)
Frame = +2
Query: 37 LVYLDHAGATLYSELQMESV---FKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVLDYC 93
LVY+D+A + ++++ ++ +NV+ H S A D AR++V +
Sbjct: 335 LVYIDNAATSQKPAAVVKALQDYYEGYNSNVHRGIHYLS---AKATDEYESARRKVAAFI 505
Query: 94 NASPEDYKCIFTSGATAALKLVGEAFPWSCNSN------FMYTMENHNSVLGIREYALGQ 147
NAS + + +FT AT A+ LV A+ W ++ + E+H++++ + A
Sbjct: 506 NAS-DSREIVFTRNATEAINLV--AYSWGLSNLKQGDEIVLTVAEHHSAIVPWQLVAQKV 676
Query: 148 GAAAIAVDI-EDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAK 206
GA V++ +D P I+ K+ ++ K+ + + + +R D+A
Sbjct: 677 GAVLKFVNLNQDEIPDID------KLKEVLSKKTKIVAVHHVSNVLASVLPIR---DIAH 829
Query: 207 IIKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLG 266
+K VL+DA + M D+ DF+ S +K+ G PTG+G
Sbjct: 830 WAHGVGAK-------------VLVDACQSVPHMVGDVQNLDADFLVASSHKMCG-PTGIG 967
Query: 267 ALVVRND 273
L + D
Sbjct: 968 FLYGKMD 988
>TC91175 weakly similar to PIR|T05129|T05129 hypothetical protein F7H19.160
- Arabidopsis thaliana, partial (18%)
Length = 981
Score = 42.7 bits (99), Expect = 5e-04
Identities = 56/258 (21%), Positives = 95/258 (36%), Gaps = 30/258 (11%)
Frame = +3
Query: 5 EFLKEFGEHYGYPNAARTIDQIRATEFNRLQDLVY--LDHAGATLYSELQMES------- 55
E EF + Y + ID +RA E+ L L LD+ G L+S Q +
Sbjct: 210 ESFTEFIKVYPQFSETEKIDSLRAKEYYHLSFLNQSCLDYIGIGLFSYYQRQQHDASKTQ 389
Query: 56 VFKDLTTNVYGNPHSQSD------------------SSAATHDIVRDARQQVLDYCNASP 97
T+ + +P SD + R++++ + N S
Sbjct: 390 FSSPSTSTPFQSPQQYSDIPFFSISYKTGNLKTLLLHGGKESEFESAMRKRIMKFLNISE 569
Query: 98 EDYKCIFTSGATAALKLVGEAFPW-SCNSNFMYTMENHNSVLGIREYALGQGAAAIAVDI 156
DY +FT+ T+A KLV +++P+ SC +V + + +GA +++ +
Sbjct: 570 NDYFMVFTANRTSAFKLVADSYPFQSCKKLLTVYDYESEAVEAMISTSENRGAKSMSAEF 749
Query: 157 EDVHPRIEGEKFPTKISLHQEQR--RKVTGLQEEEPMECNFSGLRFDLDLAKIIKEDSSK 214
RI+ K I ++ +K GL P+ +G R+
Sbjct: 750 SWPRLRIQSTKLKKMIVSDNSKKKIKKKNGL-FVFPLHSRVTGARY-------------P 887
Query: 215 ILGASVCKKGRWLVLIDA 232
L + W VLIDA
Sbjct: 888 YLWMRTAPENGWHVLIDA 941
>TC81388 weakly similar to PIR|T05129|T05129 hypothetical protein F7H19.160
- Arabidopsis thaliana, partial (18%)
Length = 1308
Score = 37.0 bits (84), Expect = 0.028
Identities = 24/67 (35%), Positives = 35/67 (51%), Gaps = 6/67 (8%)
Frame = +3
Query: 219 SVCKKGRWLVLIDAAKGSATMPPD-----LSKYPVDFVALSFYKLFG-YPTGLGALVVRN 272
S+ K+ W L+DA A P D LS + DF+ S YK+FG P+G G L ++
Sbjct: 897 SIAKENGWNGLVDAC---ALGPKDMDGFGLSLFQPDFLICSCYKVFGENPSGFGCLFIKK 1067
Query: 273 DAAKLLK 279
A +L+
Sbjct: 1068SATSILE 1088
Score = 35.8 bits (81), Expect = 0.063
Identities = 14/37 (37%), Positives = 25/37 (66%)
Frame = +1
Query: 86 RQQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPWS 122
R+++++Y N S DY +FT+ T+A KLV + + +S
Sbjct: 538 RKRIMNYLNISENDYFMVFTANRTSAFKLVADCYNFS 648
>TC88865 similar to PIR|H87729|H87729 protein Y23H5A.7 [imported] -
Caenorhabditis elegans, partial (1%)
Length = 869
Score = 33.1 bits (74), Expect = 0.41
Identities = 14/44 (31%), Positives = 26/44 (58%)
Frame = +2
Query: 85 ARQQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPWSCNSNFM 128
A+ +VL +C ++Y +FT A+ LVGE++P+ + +M
Sbjct: 668 AQNKVLKHCGLPDDEYLVLFTPSYKDAMMLVGESYPFVKGNYYM 799
>TC87015
Length = 1852
Score = 32.7 bits (73), Expect = 0.53
Identities = 20/83 (24%), Positives = 42/83 (50%)
Frame = +1
Query: 85 ARQQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPWSCNSNFMYTMENHNSVLGIREYA 144
A+ +VL + + ++Y +FT A+ LVGE++P+ + +M ++ I+E+A
Sbjct: 955 AQNKVLKHYGLTDDEYLVLFTPSYKDAMMLVGESYPFIKGNYYMTILDQEEDF--IKEFA 1128
Query: 145 LGQGAAAIAVDIEDVHPRIEGEK 167
+ + I + RI+G +
Sbjct: 1129 CFKESKVIPAPKTWLDLRIKGSQ 1197
>BG451334 weakly similar to GP|9755390|gb| F17F8.22 {Arabidopsis thaliana},
partial (48%)
Length = 653
Score = 30.8 bits (68), Expect = 2.0
Identities = 33/153 (21%), Positives = 62/153 (39%), Gaps = 10/153 (6%)
Frame = +3
Query: 524 TMGSLKHDREWILKSLSGEILTLKRVPEMGLI----------SSFIDLSQGMLFVESPHC 573
T L+ DR+W++ + G T + P++ L+ + + + +++P
Sbjct: 123 TPSGLRWDRQWVVVNSKGRACTQRVEPKLALVEVELPPEAFDEHWEPTTDSFMVLKAPG- 299
Query: 574 KERLQIRLQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDF 633
E L++ L + DI + + E WFS + P L+R++ +S
Sbjct: 300 MEPLKVFLNKQY--EVADDITVWEWTGSAWDEGAEAAQWFSDYLGNPTKLVRFNTASEVR 473
Query: 634 VLDRTKDIVTCKDTNSAVSFANEGQFLLVSEES 666
+D D V + F + FLLVS+ S
Sbjct: 474 KVD--PDYV---EGQQQTFFTDGYPFLLVSQAS 557
>TC77300 similar to PIR|T48643|T48643 cinnamoyl CoA reductase-like protein -
Arabidopsis thaliana, partial (63%)
Length = 1559
Score = 29.3 bits (64), Expect = 5.9
Identities = 17/75 (22%), Positives = 35/75 (46%)
Frame = +1
Query: 62 TNVYGNPHSQSDSSAATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPW 121
T+ + +P S + + +I + + V++ C +P +C+FTS A + +
Sbjct: 610 TSAFTDPAGLSGYTKSMAEIEVNVTENVMEACARTPSIKRCVFTSSLAACI------WQD 771
Query: 122 SCNSNFMYTMENHNS 136
+ NS+ + NH S
Sbjct: 772 NVNSDLTTPIINHGS 816
>TC86447 weakly similar to GP|19911585|dbj|BAB86896. syringolide-induced
protein 13-1-1 {Glycine max}, partial (36%)
Length = 1223
Score = 29.3 bits (64), Expect = 5.9
Identities = 14/33 (42%), Positives = 21/33 (63%)
Frame = +1
Query: 87 QQVLDYCNASPEDYKCIFTSGATAALKLVGEAF 119
Q++ + N S + KCI +GAT+AL L +AF
Sbjct: 694 QKMEKWSNESERNKKCIVENGATSALALAFDAF 792
>TC89759 similar to GP|13430174|gb|AAK25754.1 putative chaperon P13.9
{Castanea sativa}, partial (54%)
Length = 646
Score = 29.3 bits (64), Expect = 5.9
Identities = 17/39 (43%), Positives = 24/39 (60%), Gaps = 3/39 (7%)
Frame = +3
Query: 422 EAGHVCW---DDQDIISGKPIGAVRVSFGYMSTFEDAKK 457
+AG +CW +DI+ G GA + G+MSTF+D KK
Sbjct: 354 KAGGLCWLCRGKKDILCGSCNGAGFIG-GFMSTFDD*KK 467
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,840,096
Number of Sequences: 36976
Number of extensions: 312061
Number of successful extensions: 1212
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1197
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1209
length of query: 764
length of database: 9,014,727
effective HSP length: 104
effective length of query: 660
effective length of database: 5,169,223
effective search space: 3411687180
effective search space used: 3411687180
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148651.4


