

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.11 - phase: 0
(395 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
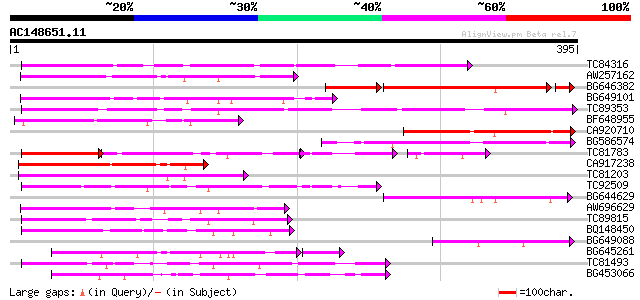
Score E
Sequences producing significant alignments: (bits) Value
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 144 8e-35
AW257162 136 2e-32
BG646382 123 2e-32
BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidops... 130 9e-31
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 122 2e-28
BF648955 similar to GP|18043667|gb| Unknown (protein for IMAGE:3... 119 3e-27
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 117 1e-26
BG586574 117 1e-26
TC81783 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 60 5e-26
CA917238 weakly similar to PIR|A84538|A845 hypothetical protein ... 110 1e-24
TC81203 weakly similar to GP|10177464|dbj|BAB10855. gb|AAD21700.... 107 8e-24
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 97 1e-20
BG644629 89 4e-18
AW696629 86 2e-17
TC89815 85 6e-17
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 84 1e-16
BG649088 81 6e-16
BG645261 homologue to PIR|H90254|H902 sulfate ABC transporter p... 79 9e-16
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 79 2e-15
BG453066 78 7e-15
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 144 bits (362), Expect = 8e-35
Identities = 97/315 (30%), Positives = 152/315 (47%), Gaps = 1/315 (0%)
Frame = +3
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNF 68
LP EL+ IL LPV+ L+RF+CV K + TL SD HF H S P L+ +
Sbjct: 42 LPEELIVIILLRLPVRSLLRFKCVCKSWKTLFSDTHFANNHFLISTVYPQLVACESVSAY 221
Query: 69 NSFDENVLNLPISLLLENSLSTV-PYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTS 127
+++ + PI LLENS +TV P + H + GSCNG +CL +
Sbjct: 222 RTWE--IKTYPIESLLENSSTTVIPV-------SNTGHQRYTILGSCNGFLCLY----DN 362
Query: 128 HGSRLCLWNPATRTKSEFDLASQECFVFAFGYDNLNGNYKVIAFDIKVKSGNARSVVKVF 187
+ + LWNP+ KS+ + FGYD +N YK++A VK+ + + ++
Sbjct: 363 YQRCVRLWNPSINLKSKSSPTIDRFIYYGFGYDQVNHKYKLLA----VKAFSRITETMIY 530
Query: 188 SMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFGLDYFHLNYSSITPEKY 247
+ +N +N++ P Y G + S T+NW+ + +
Sbjct: 531 TFGENSCKNVE-VKDFPRYPPNRKHLGKFVSGTLNWIVDE---------------RDGRA 662
Query: 248 VILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQ 307
ILS D+ ETY Q+LLP+ + + + P L VL +C+C + +T + +W MK +GV
Sbjct: 663 TILSFDIEKETYRQVLLPQ--HGYAVYSPGLYVLSNCICVCTSFLDTRWQLWMMKKYGVA 836
Query: 308 SSWIQLFKITYQNFF 322
SW +L I ++N F
Sbjct: 837 ESWTKLMSIPHENLF 881
>AW257162
Length = 728
Score = 136 bits (342), Expect = 2e-32
Identities = 79/200 (39%), Positives = 114/200 (56%), Gaps = 6/200 (3%)
Frame = +1
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHN 67
V P +++ +ILS L VK LM+ +CV+K +NTLISD +F++MHL SAR+ +++
Sbjct: 124 VFPDDIIAEILSWLTVKTLMKMKCVSKSWNTLISDSNFVKMHLNRSARHSQSYLVSEHRG 303
Query: 68 FNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICL--CL-DI 124
D N + + L+ T+P DPYY+L ++ CP + GSCNGL+CL C+ D+
Sbjct: 304 ----DYNFVPFSVRGLMNGRSITLPKDPYYQLIEKD--CPGV-VGSCNGLVCLSGCVADV 462
Query: 125 DTSHGSRLCLWNPATRTKSE---FDLASQECFVFAFGYDNLNGNYKVIAFDIKVKSGNAR 181
+ L +WNPATRT S+ F +C+ F FGYDN YKV+A
Sbjct: 463 EEFEEMWLRIWNPATRTISDKLYFSANRLQCWEFMFGYDNTTQTYKVVAL---YPDSEMT 633
Query: 182 SVVKVFSMRDNCWRNIQCFP 201
+ V + R+N WRNIQ FP
Sbjct: 634 TKVGIICFRNNIWRNIQSFP 693
>BG646382
Length = 603
Score = 123 bits (308), Expect(3) = 2e-32
Identities = 64/124 (51%), Positives = 79/124 (63%), Gaps = 7/124 (5%)
Frame = +3
Query: 261 QLLLPRGFNKVSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITYQN 320
Q LLP GF +V +P L VLMDCLCF HD++ T FVIWQMK+FG Q SW QLF+I Y N
Sbjct: 186 QFLLPAGFKEVPCVEPSLRVLMDCLCFSHDFKRTEFVIWQMKEFGSQESWTQLFRIKYIN 365
Query: 321 FFSYYCDFARESKWLDF-------LPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIG 373
+ L + LPL LS+NGDTLIL+N++E IYN+RD RVEK
Sbjct: 366 LQIHNLPINDNLDLLGYMECNIPLLPLYLSENGDTLILSNDEEEGVIIYNQRDKRVEKTR 545
Query: 374 ITNK 377
I+N+
Sbjct: 546 ISNE 557
Score = 30.4 bits (67), Expect(3) = 2e-32
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 2/41 (4%)
Frame = +2
Query: 221 VNWLALQ-DYFGLDYFHLNYSSIT-PEKYVILSLDLSTETY 259
+ WLAL D+ +D + + I E+ VI+SLDLSTETY
Sbjct: 59 LTWLALSGDFVSIDCGETSKAYIPLVEQLVIVSLDLSTETY 181
Score = 23.9 bits (50), Expect(3) = 2e-32
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +1
Query: 381 WFEAWDYVESLVS 393
W A+DYVESLVS
Sbjct: 565 WPSAFDYVESLVS 603
>BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 799
Score = 130 bits (327), Expect = 9e-31
Identities = 89/234 (38%), Positives = 129/234 (55%), Gaps = 13/234 (5%)
Frame = +1
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHN 67
V+PS+L+ IL+ LPVK + + + V+K +NTLI+ P FI++HL S++NPN ++ +
Sbjct: 82 VIPSDLIAIILTFLPVKTITQLKLVSKSWNTLITSPSFIKIHLNQSSQNPNFILTPSRKQ 261
Query: 68 FNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCL--DID 125
++ NVL++PI LL +TV D Y+ + N + H + GSCNGL+CL +
Sbjct: 262 YSI--NNVLSVPIPRLLTG--NTVSGDTYHNILNNDHH--FRVVGSCNGLLCLLFKSEFI 423
Query: 126 TSHGSRLCLWNPATRTKSE---FDLASQECF----VFAFGYDNLNGNYKVIAFDIKVKSG 178
T R +WNPATRT SE F + F F FG D L G YK++A
Sbjct: 424 THLKFRFRIWNPATRTISEELGFFRKYKPLFGGVSRFTFGCDYLRGTYKLVALHTVEDGD 603
Query: 179 NARSVVKVFSM----RDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQD 228
RS V+VF++ D CWRNI FV +GV+ S T NWL+L++
Sbjct: 604 VMRSNVRVFNLGNDDSDKCWRNIP-------NPFVCA-DGVHLSGTGNWLSLRE 741
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 122 bits (307), Expect = 2e-28
Identities = 113/404 (27%), Positives = 178/404 (43%), Gaps = 17/404 (4%)
Frame = +3
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNF 68
+P E++ +IL LPV+ L++FRCV K + TLISDP F + H+ S P L+ + F
Sbjct: 105 MPEEIIVEILLRLPVRSLLQFRCVCKLWKTLISDPQFAKKHVSISTAYPQLVSV-----F 269
Query: 69 NSFDE-NVLNLPISLLLEN-SLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDT 126
S + N+++ P+ LL+N S V + + + GSCNGL+CL
Sbjct: 270 VSIAKCNLVSYPLKPLLDNPSAHRVEPADFEMIHTTS----MTIIGSCNGLLCL------ 419
Query: 127 SHGSRLCLWNPATRTKSE-------FDLASQECFVF-AFGYDNLNGNYKVIAFDIKVKS- 177
S + LWNP+ + KS+ FD + F++ FGYD +N YKV+A +
Sbjct: 420 SDFYQFTLWNPSIKLKSKPSPTIIAFDSFDSKRFLYRGFGYDQVNDRYKVLAVVQNCYNL 599
Query: 178 GNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQN-GVYFSSTVNWLALQDYFGLDYFH 236
++++ F +D W IQ FP P + G + S +NW+
Sbjct: 600 DETKTLIYTFGGKD--WTTIQKFPCDPSRCDLGRLGVGKFVSGNLNWIV----------- 740
Query: 237 LNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDY-EETY 295
K VI+ D+ ETY ++ LP+ + L V + + D+ +T+
Sbjct: 741 --------SKKVIVFFDIEKETYGEMSLPQDYGD---KNTVLYVSSNRIYVSFDHSNKTH 887
Query: 296 FVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARESKWLDFLPLCLSK----NGDTLIL 351
+V+W MK++GV SW +L I + K P CLS + ++L
Sbjct: 888 WVVWMMKEYGVVESWTKLMIIP-------------QDKLTSPGPYCLSDALFISEHGVLL 1028
Query: 352 ANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVSTH 395
Q ++ +YN +D T Y ESLVS H
Sbjct: 1029MRPQHSKLSVYNLNNDGGLDYCTTISGQFARYLHIYNESLVSPH 1160
>BF648955 similar to GP|18043667|gb| Unknown (protein for IMAGE:3661715) {Mus
musculus}, partial (3%)
Length = 514
Score = 119 bits (297), Expect = 3e-27
Identities = 76/171 (44%), Positives = 102/171 (59%), Gaps = 11/171 (6%)
Frame = +3
Query: 4 RHQSV-----LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPN 58
RH+S+ LPSEL+ +I+S LPVK LM+FRCV+KFY TLISDP+F+QMHL+ SARNP+
Sbjct: 27 RHRSMASAAFLPSELIVEIISWLPVKYLMQFRCVSKFYKTLISDPYFVQMHLEKSARNPH 206
Query: 59 LMVIARQHNFNSFDENVLNLPISLLLENSLSTVPYD--PYYRLKNENPHCPWLFAGSCNG 116
L + Q + D +++ L +S LL N +T P+ +Y GSCNG
Sbjct: 207 L-ALMWQDDLLREDGSIIFLSVSRLLGNKYTTPPFQSGTFYHS---------CIIGSCNG 356
Query: 117 LICLCLDI---DTSHGSRLCLWNPATRTK-SEFDLASQECFVFAFGYDNLN 163
L+CL +D D + L WNPATRTK + F F+FGYD L+
Sbjct: 357 LLCL-VDFHCPDYYYYYYLYFWNPATRTKFXNILITLSXDFKFSFGYDTLS 506
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 117 bits (292), Expect = 1e-26
Identities = 61/123 (49%), Positives = 84/123 (67%), Gaps = 3/123 (2%)
Frame = -2
Query: 275 QPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARESKW 334
+P L VL+DCLCF HD +T FVIWQMK+FGV+ SW +LFKI Y + + + ++
Sbjct: 764 EPYLRVLLDCLCFLHDLGKTEFVIWQMKEFGVRESWTRLFKIPYVDL--QMLNLPIDVQY 591
Query: 335 LD---FLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESL 391
L+ LPL +SKNGD +IL N +++ IYN+RD RV++ I+N+ + WF A DYVESL
Sbjct: 590 LNEYPMLPLYISKNGDKVILTNEKDDATIIYNKRDKRVDRARISNE-IHWFSAMDYVESL 414
Query: 392 VST 394
V T
Sbjct: 413 VPT 405
>BG586574
Length = 732
Score = 117 bits (292), Expect = 1e-26
Identities = 76/185 (41%), Positives = 100/185 (53%), Gaps = 8/185 (4%)
Frame = +1
Query: 218 SSTVNWLA-LQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLP-------RGFN 269
+ST+NWL +QD + P+ VI+SLDL TETYTQ L P
Sbjct: 4 NSTLNWLGFIQD-----------GDLAPQ-LVIISLDLGTETYTQFLPPPISLDLSHVLQ 147
Query: 270 KVSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFA 329
KVS +P +++LMD LCF HD ET FVIW+M FG + SW QL KI+Y
Sbjct: 148 KVSHAKPGVSMLMDSLCFYHDLNETDFVIWKMTKFGDEKSWAQLLKISYHKL-------- 303
Query: 330 RESKWLDFLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVE 389
+ + L NGDTL+ ++Q+ A +YN R++RV K + NK + WF YVE
Sbjct: 304 KMNLKPGISMFNLYVNGDTLVFVDDQKERAILYNWRNNRVVKTRV-NKKICWFSINHYVE 480
Query: 390 SLVST 394
SLVST
Sbjct: 481 SLVST 495
>TC81783 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (6%)
Length = 1225
Score = 59.7 bits (143), Expect(4) = 5e-26
Identities = 28/57 (49%), Positives = 43/57 (75%)
Frame = +2
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQ 65
LP +L+ +IL LPVK L++ RC+ KF+N+LISDP+F + H + SA++ LMV +R+
Sbjct: 134 LPFDLLPEILCRLPVKLLIQLRCLCKFFNSLISDPNFAKKHFQFSAKHHRLMVTSRK 304
Score = 40.4 bits (93), Expect(4) = 5e-26
Identities = 22/63 (34%), Positives = 37/63 (57%), Gaps = 5/63 (7%)
Frame = +2
Query: 278 LAVLM--DCLCFGHDYEETYFVIWQMKDFGVQSSWIQLF---KITYQNFFSYYCDFARES 332
L+VLM DCLC ++ + +F +W MK++G Q SW +L+ K+ ++ F +Y + E
Sbjct: 863 LSVLMLRDCLCV-YEASDLFFNVWIMKEYGNQESWTKLYSVPKMQHRRFKAYTISYIYED 1039
Query: 333 KWL 335
L
Sbjct: 1040DQL 1048
Score = 40.0 bits (92), Expect(4) = 5e-26
Identities = 39/150 (26%), Positives = 65/150 (43%), Gaps = 8/150 (5%)
Frame = +3
Query: 65 QHNFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNEN-PHCPWLFAGSCNGLICLCLD 123
+H+F D + P S++ DP + L N + + SC+G+ C L+
Sbjct: 309 RHDFVLHDSPI---PSVFSTSTSVTQTQLDPPFTLARRNYVNATAIAMCSCDGIFCGELN 479
Query: 124 IDTSHGSRLCLWNPATRTKSEFDLASQ-------ECFVFAFGYDNLNGNYKVIAFDIKVK 176
+ +WNP+ R +F L + F +FGYD+ NYKV+A K +
Sbjct: 480 LGYCF-----VWNPSIR---KFKLLPPYKNPLEGDPFSISFGYDHFIDNYKVVAISSKYE 635
Query: 177 SGNARSVVKVFSMRDNCWRNIQCFPVLPLY 206
V V ++ + WR I+ P+L L+
Sbjct: 636 -------VFVNALGTDYWRRIREHPLLLLH 704
Score = 35.0 bits (79), Expect(4) = 5e-26
Identities = 22/68 (32%), Positives = 32/68 (46%)
Frame = +1
Query: 203 LPLYMFVSTQNGVYFSSTVNWLALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQL 262
+P Y ++ Q G++ TVNW+A S + ILSLDL E+Y +L
Sbjct: 685 IPYYYYI-LQPGIFVGGTVNWMA---------------SDIADLLFILSLDLEKESYQKL 816
Query: 263 LLPRGFNK 270
LP N+
Sbjct: 817 FLPDSENE 840
>CA917238 weakly similar to PIR|A84538|A845 hypothetical protein At2g16220
[imported] - Arabidopsis thaliana, partial (6%)
Length = 603
Score = 110 bits (274), Expect = 1e-24
Identities = 60/134 (44%), Positives = 86/134 (63%), Gaps = 2/134 (1%)
Frame = +2
Query: 7 SVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH 66
SVLP EL+T++LS VK LMR RCV++++N++ISDP F+++H+K SARN +L + +
Sbjct: 140 SVLPDELITEVLSRGDVKSLMRMRCVSEYWNSMISDPRFVKLHMKRSARNAHLTLSLCKS 319
Query: 67 NFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLC--LDI 124
+ D NV+ P+ L+EN L T+P DPYYRLK++ C ++ GSCNG +CL I
Sbjct: 320 GIDG-DNNVVPYPVRGLIENGLITLPSDPYYRLKDK--ECQYVI-GSCNGWLCLLGFSSI 487
Query: 125 DTSHGSRLCLWNPA 138
WNPA
Sbjct: 488 GAYRHIWFRFWNPA 529
>TC81203 weakly similar to GP|10177464|dbj|BAB10855.
gb|AAD21700.1~gene_id:MQB2.18~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 715
Score = 107 bits (267), Expect = 8e-24
Identities = 68/174 (39%), Positives = 96/174 (55%), Gaps = 14/174 (8%)
Frame = +1
Query: 7 SVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLIS-DPHFIQMHLKNSARNPNLMVIARQ 65
S+L EL+ ILS LPVK LM+F+CV K + TLIS DP F ++HL+ S RN +L +++ +
Sbjct: 193 SLLLDELIVDILSRLPVKTLMQFKCVCKSWKTLISHDPSFAKLHLQRSPRNTHLTLVSDR 372
Query: 66 HNFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKN-ENPHCPW---------LFAGSCN 115
+ + + +V+ P+S L+E L+ P++PY+ L+N P P+ + GSCN
Sbjct: 373 SSDDESNFSVVPFPVSHLIEAPLTITPFEPYHLLRNVAIPDDPYYVLGNMDCCIIIGSCN 552
Query: 116 GLICL---CLDIDTSHGSRLCLWNPATRTKSEFDLASQECFVFAFGYDNLNGNY 166
GL CL L + WNPAT T SE E F FGYD N Y
Sbjct: 553 GLYCLRCYSLIYEEEEHDWFRFWNPATNTLSEELGCLNEFFRLTFGYDISNDTY 714
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 96.7 bits (239), Expect = 1e-20
Identities = 83/259 (32%), Positives = 120/259 (46%), Gaps = 8/259 (3%)
Frame = +3
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNF 68
LP EL+ +IL LPVK L + RC+ K +N+LISDP F + HL +S+ P+ +++ +
Sbjct: 174 LPFELVAEILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHL-HSSTTPHHLILRSNNGS 350
Query: 69 NSFDENVLNLPISLLLENSLSTVPYD--PYYRLKNENPHCPWLFAGSCNGLICLCLDIDT 126
F ++ PI +L S VP Y E P+ + SC+G+ICL D
Sbjct: 351 GRF--ALIVSPIQSVLSTSTVPVPQTQLTYPTCLTEEFASPYEWC-SCDGIICLTTDY-- 515
Query: 127 SHGSRLCLWNP------ATRTKSEFDLASQECFVFAFGYDNLNGNYKVIAFDIKVKSGNA 180
S LWNP L +F FGYD NYKV A VK
Sbjct: 516 ---SSAVLWNPFINKFKTLPPLKYISLKRSPSCLFTFGYDPFADNYKVFAITFCVK---- 674
Query: 181 RSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFGLDYFHLNYS 240
R+ V+V +M + WR I+ F P + F+ +G++ + V+WL Y G
Sbjct: 675 RTTVEVHTMGTSSWRRIEDF---PSWSFI-PDSGIFVAGYVHWLT---YDG--------- 806
Query: 241 SITPEKYVILSLDLSTETY 259
+ I+SLDL E+Y
Sbjct: 807 --PGSQREIVSLDLEDESY 857
>BG644629
Length = 546
Score = 88.6 bits (218), Expect = 4e-18
Identities = 55/148 (37%), Positives = 80/148 (53%), Gaps = 16/148 (10%)
Frame = -3
Query: 261 QLLLPRGFNKVSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITYQN 320
Q LPR F+++ P +AV LC + Y+ET F+IWQMK+ SW Q KI YQN
Sbjct: 544 QYRLPRDFHEMPSALPIVAV*GGFLCCSYFYKETDFLIWQMKELANDESWTQFLKIRYQN 365
Query: 321 F---FSYYCD--FARESKWLDF--LPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIG 373
Y+ D F ++ F +PL LS+ DTL+L +QE+ +YN +++RV++
Sbjct: 364 LQINHDYFSDHEFIHDTVKYHFQLVPLLLSEEADTLVLKTSQESHPILYNWKENRVQRTK 185
Query: 374 IT---------NKNMLWFEAWDYVESLV 392
+T ++LW A YVESLV
Sbjct: 184 VTTSTTVTDDSRNSILWCSAKVYVESLV 101
>AW696629
Length = 663
Score = 86.3 bits (212), Expect = 2e-17
Identities = 62/200 (31%), Positives = 102/200 (51%), Gaps = 12/200 (6%)
Frame = +2
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHN 67
+LPS+L+ +ILS LPVK L+RF V+K + +LI DP+F ++HL+ S +N ++++ A
Sbjct: 62 ILPSDLIMQILSWLPVKLLIRFTSVSKHWKSLILDPNFAKLHLQKSPKNTHMILTALDDE 241
Query: 68 FNSFDENVLNLPI-SLLLENSLSTVPYDPYYRLKNENPHC----PWLFAGSCNGLICLCL 122
+++ V P+ SLLLE S +E C + GS NGL+CL +
Sbjct: 242 DDTW--VVTPYPVRSLLLEQS----------SFSDEECCCFDYHSYFIVGSTNGLVCLAV 385
Query: 123 DIDTSHGSR---LCLWNPATRTKSE----FDLASQECFVFAFGYDNLNGNYKVIAFDIKV 175
+ + + WNP+ R +S+ ++ FGYD+LN YK +A +
Sbjct: 386 EKSLENRKYELFIKFWNPSLRLRSKKAPSLNIGLYGTARLGFGYDDLNDTYKAVA--VFW 559
Query: 176 KSGNARSVVKVFSMRDNCWR 195
+ +V M D+CW+
Sbjct: 560 DHTTHKMEGRVHCMGDSCWK 619
>TC89815
Length = 807
Score = 84.7 bits (208), Expect = 6e-17
Identities = 62/202 (30%), Positives = 96/202 (46%), Gaps = 13/202 (6%)
Frame = +1
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNF 68
+P E+ ILSLLP PL+RFR +K ++I F +HLKNS N + +HN
Sbjct: 46 IPPEIFIDILSLLPPHPLLRFRSTSKSLKSIIDSHTFTNLHLKNS----NNFYLIIRHNA 213
Query: 69 NSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTSH 128
N + + NL + L + L + Y L GSCNGLIC+ ++
Sbjct: 214 NLYQLDFPNLTPPIPLNHPL--MSYSNRITL-----------FGSCNGLICI-----SNI 339
Query: 129 GSRLCLWNP-----------ATRTKSEFDLASQECFVFAFGYDNLNGNYKV--IAFDIKV 175
+ WNP T +SE D V FGYD+ G+YK+ I++ + +
Sbjct: 340 ADDIAFWNPNIRKHRIIPYLPTTPRSESDTTLFAARVHGFGYDSFAGDYKLVRISYFVDL 519
Query: 176 KSGNARSVVKVFSMRDNCWRNI 197
++ + S V+VFS++ N W+ +
Sbjct: 520 QNRSFDSQVRVFSLKMNSWKEL 585
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 83.6 bits (205), Expect = 1e-16
Identities = 65/200 (32%), Positives = 102/200 (50%), Gaps = 10/200 (5%)
Frame = +3
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLK-NSARNPNLMVIARQHN 67
LP E+ +ILS LPVK LM+F+CV K + + IS P F++ HL+ ++ R+ L+ ++
Sbjct: 3 LPFEIQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKHLRVSNTRHLFLLTFSKL-- 176
Query: 68 FNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTS 127
S + + + P+S + T Y L N + GSC+G++C+ ++
Sbjct: 177 --SPELVIKSYPLSSVFTEMTPTFT-QLEYPLNNRDESDS--MVGSCHGILCIQCNL--- 332
Query: 128 HGSRLCLWNPATR--TKSEFDLASQECFV---FAFGYDNLNGNYKVIAFDIKVKSGNA-- 180
S LWNP+ R TK Q F+ +AFGYD+ + YKV+A N
Sbjct: 333 --SFPVLWNPSIRKFTKLPSFEFPQNKFINPTYAFGYDHSSDTYKVVAVFCTSNIDNGVY 506
Query: 181 --RSVVKVFSMRDNCWRNIQ 198
+++V V +M NCWR IQ
Sbjct: 507 QLKTLVNVHTMGTNCWRRIQ 566
>BG649088
Length = 683
Score = 81.3 bits (199), Expect = 6e-16
Identities = 46/107 (42%), Positives = 62/107 (56%), Gaps = 8/107 (7%)
Frame = -3
Query: 295 YFVIWQMKDFGVQSSWIQLFKITYQNFFSYY--CDFARESKWLDFLPLCLSKNGDTLILA 352
+FV+W M ++GVQ SW Q KI++Q+ Y D L P LS++ +TLI+A
Sbjct: 675 HFVLWLMMEYGVQDSWTQFLKISFQDLQIDYGISDSLDYGSQLYLYPFYLSESDNTLIMA 496
Query: 353 NNQE------NEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVS 393
+NQ+ N A IYN RD VE+I + +LWF DYVESLVS
Sbjct: 495 SNQQGYDGYDNHAIIYNWRDKTVEQITSVDNEILWFHTKDYVESLVS 355
>BG645261 homologue to PIR|H90254|H902 sulfate ABC transporter permease
protein SSO1032 [imported] - Sulfolobus solfataricus,
partial (4%)
Length = 771
Score = 78.6 bits (192), Expect(2) = 9e-16
Identities = 64/197 (32%), Positives = 100/197 (50%), Gaps = 23/197 (11%)
Frame = +1
Query: 30 RCVNKFYNTLISDPHFIQMHLKNSARNPNLMVI--ARQHNFNSFDENVLNLPISLLLENS 87
R +N + ++I P F+++HL S++ P+L + AR + ++ + LLEN
Sbjct: 112 RFINILWKSIIDHPTFVKLHLNRSSQKPDLTFVSSARITLWTLESTCTVSFTVFRLLENP 291
Query: 88 ---LSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLC-LDIDTSHGSR---LCLWNPATR 140
++ DPYY+LK+++ C + GSCNGL+CL L I+ S G + WNPA+R
Sbjct: 292 PVIINLSDDDPYYQLKDKD--CVHII-GSCNGLLCLFGLKINDSCGHMDISIRFWNPASR 462
Query: 141 TKSEF-----DLASQE--CFV-------FAFGYDNLNGNYKVIAFDIKVKSGNARSVVKV 186
S+ D + + CF F FGY N + YKV+ F + S +V
Sbjct: 463 KISKKVGYCGDGVNNDPSCFPGKGDLLKFVFGYVNSSDTYKVVYFIVGTTSA------RV 624
Query: 187 FSMRDNCWRNIQCFPVL 203
FS+ DN WR I+ PV+
Sbjct: 625 FSLGDNVWRKIENSPVV 675
Score = 22.3 bits (46), Expect(2) = 9e-16
Identities = 10/29 (34%), Positives = 15/29 (51%)
Frame = +2
Query: 205 LYMFVSTQNGVYFSSTVNWLALQDYFGLD 233
L++F+ Q V WL +Q+Y G D
Sbjct: 668 LWLFIMCQGLSI*VVVVFWLTIQNYHGYD 754
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 728
Score = 79.3 bits (194), Expect = 2e-15
Identities = 70/281 (24%), Positives = 124/281 (43%), Gaps = 24/281 (8%)
Frame = +1
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH-- 66
LP+E+ T+ILS +P KPL+R R K++ LI FI +HL S + ++I RQH
Sbjct: 16 LPTEVTTEILSRVPAKPLLRLRSTCKWWRNLIDSTDFIFLHLSKSR---DSVIILRQHSR 186
Query: 67 ----NFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCL 122
+ NS D V L L+ ++ R+K GSCNGL+C+C
Sbjct: 187 LYELDLNSMD-RVKELDHPLMCYSN----------RIK---------VLGSCNGLLCICN 306
Query: 123 DIDTSHGSRLCLWNPATR---------------TKSEFDLASQECFVFAFGYDNLNGNYK 167
D + WNP R ++ V+ FGYD+ +YK
Sbjct: 307 IADD-----IAFWNPTIRKHRIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDYK 471
Query: 168 VIAFD--IKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLA 225
+++ + + + + S V +++M + W + P +P + + GV+ S ++W+
Sbjct: 472 LVSISNFVDLHNRSYDSHVTIYTMGSDVWMPL---PGVPYALCCARPMGVFVSGALHWVV 642
Query: 226 LQDYFGLDYFHLNYSSITPE-KYVILSLDLSTETYTQLLLP 265
+ ++ P+ + +I++ DL E + ++ LP
Sbjct: 643 PR-------------ALEPDSRDLIVAFDLRFEVFREVALP 726
>BG453066
Length = 655
Score = 77.8 bits (190), Expect = 7e-15
Identities = 75/250 (30%), Positives = 116/250 (46%), Gaps = 14/250 (5%)
Frame = +1
Query: 30 RCVNKFYNTLISDPHFIQMHLKNSARNPNLMV--IARQHNFNSFDENVLNL--PISLLLE 85
RC+ KF+N+LISDP F++ HL++S + +LM+ I Q F +D + +L +++ +
Sbjct: 4 RCLCKFFNSLISDPKFVKKHLQSSTKRRHLMLTTIDHQQQFVMYDSPIPSLFSTSTIVAQ 183
Query: 86 NSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTSHGSRLCLWNPATRT-KSE 144
L D Y +K C SC+G I L + TS+ LWNP+ R K
Sbjct: 184 TQLYPPNGDTYASVK-----C------SCDG-IFLGMFSHTSY----FLWNPSIRKFKLL 315
Query: 145 FDLASQE---------CFVFAFGYDNLNGNYKVIAFDIKVKSGNARSVVKVFSMRDNCWR 195
L +Q+ + +FGYD YKVIA ++++ V ++++ + W+
Sbjct: 316 PPLENQDKSAPFTIIVPYTISFGYDCFIDKYKVIAV-------SSKNEVFLYTLGTDYWK 474
Query: 196 NIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFGLDYFHLNYSSITPEKYVILSLDLS 255
I P Y ++G+Y S VNW Y H S Y ILSLDL
Sbjct: 475 RIDDIP----YYCTICRSGLYVSGAVNW----------YVHDESXS---SLYFILSLDLX 603
Query: 256 TETYTQLLLP 265
E+Y +L P
Sbjct: 604 NESYQELYPP 633
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.139 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,271,056
Number of Sequences: 36976
Number of extensions: 295176
Number of successful extensions: 2129
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 2020
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2046
length of query: 395
length of database: 9,014,727
effective HSP length: 98
effective length of query: 297
effective length of database: 5,391,079
effective search space: 1601150463
effective search space used: 1601150463
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148651.11


