

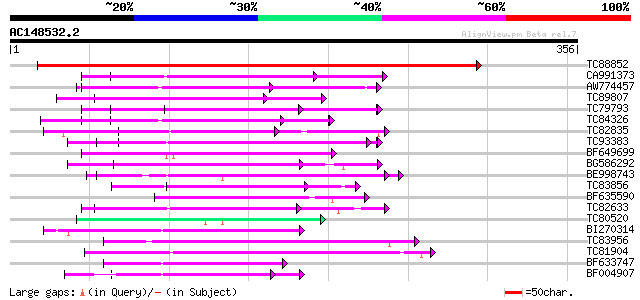
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148532.2 - phase: 0
(356 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88852 weakly similar to GP|22773243|gb|AAN06849.1 Hypothetical... 551 e-157
CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imp... 69 3e-12
AW774457 weakly similar to GP|10176973|dbj gb|AAF19552.1~gene_id... 68 5e-12
TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [i... 68 6e-12
TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Ar... 66 2e-11
TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein... 62 3e-10
TC82835 weakly similar to GP|21741712|emb|CAD41335. oj991113_30.... 60 1e-09
TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16... 59 4e-09
BF649699 similar to GP|9757911|dbj gb|AAF19552.1~gene_id:MMN10.1... 57 1e-08
BG586292 similar to PIR|T46163|T46 nodulin / glutamate-ammonia l... 55 3e-08
BE998743 similar to GP|22128591|gb| fertility restorer-like prot... 54 7e-08
TC83856 weakly similar to PIR|B96656|B96656 unknown protein 419... 54 1e-07
BF635590 weakly similar to PIR|D84778|D84 probable salt-inducibl... 53 2e-07
TC82633 similar to GP|15450347|gb|AAK96467.1 At1g20300/F14O10_8 ... 52 3e-07
TC80520 weakly similar to PIR|T02562|T02562 probable salt-induci... 52 3e-07
BI270314 similar to GP|9759030|dbj contains similarity to salt-i... 52 5e-07
TC83956 weakly similar to GP|15982931|gb|AAL09812.1 AT3g53700/F4... 52 5e-07
TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Ar... 50 1e-06
BF633747 similar to GP|10177016|db maize crp1 protein-like {Arab... 49 2e-06
BF004907 weakly similar to GP|8953393|emb putative protein {Arab... 49 2e-06
>TC88852 weakly similar to GP|22773243|gb|AAN06849.1 Hypothetical protein
{Oryza sativa (japonica cultivar-group)}, partial (36%)
Length = 1351
Score = 551 bits (1421), Expect = e-157
Identities = 278/279 (99%), Positives = 278/279 (99%)
Frame = +3
Query: 18 VLEWILEKLLKENVKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLIS 77
VLEWILEKLLKENVKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLIS
Sbjct: 249 VLEWILEKLLKENVKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLIS 428
Query: 78 ACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLG 137
ACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLG
Sbjct: 429 ACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLG 608
Query: 138 PDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEF 197
PDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEF
Sbjct: 609 PDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEF 788
Query: 198 FKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELLETITSYPIDSGVLSQIHCGIVTMY 257
FKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELLETITSYPIDSGVLSQIHCGIVTMY
Sbjct: 789 FKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELLETITSYPIDSGVLSQIHCGIVTMY 968
Query: 258 AMLDRLDEVELSVGRMLKQGMSFTSSDDVEKVICSYFRK 296
AMLD LDEVELSVGRMLKQGMSFTSSDDVEKVICSYFRK
Sbjct: 969 AMLDILDEVELSVGRMLKQGMSFTSSDDVEKVICSYFRK 1085
>CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 756
Score = 68.9 bits (167), Expect = 3e-12
Identities = 48/192 (25%), Positives = 82/192 (42%)
Frame = +2
Query: 46 KLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDF 105
K+K V +N+F M + P + +NSLI I A L + M PD
Sbjct: 167 KIKKVDEAMNLFKEMHHKHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHD-RGVPPDI 343
Query: 106 HTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEE 165
TY++ + A K+ VD +A + K G+ P++ T+ +I G E A +FE+
Sbjct: 344 ITYSSILDALCKNHQVDKAIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFED 523
Query: 166 MKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQ 225
+ + V M+ GFC++ A + DN +++ ++ +
Sbjct: 524 LLVKGYNITVNTYTVMIHGFCNKGLFDEALTLLSKMKDNSCIPDAVTYEIIIRSLFDKDE 703
Query: 226 VEKMEELLETIT 237
+K E+L E IT
Sbjct: 704 NDKAEKLREMIT 739
Score = 48.9 bits (115), Expect = 3e-06
Identities = 32/131 (24%), Positives = 55/131 (41%)
Frame = +2
Query: 64 GVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDA 123
G+KP + + SL+ ++ A S+ M S PD +YN I F K VD
Sbjct: 11 GIKPIVVTYCSLMDGYCLVKEVNKAKSILYTM-SQRGVNPDIQSYNILIDGFCKIKKVDE 187
Query: 124 MLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLK 183
+ + + PD+ T+ S+I G A ++ +EM + P++ S+L
Sbjct: 188 AMNLFKEMHHKHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSILD 367
Query: 184 GFCSQKSLCRA 194
C + +A
Sbjct: 368 ALCKNHQVDKA 400
>AW774457 weakly similar to GP|10176973|dbj
gb|AAF19552.1~gene_id:MTG13.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (13%)
Length = 650
Score = 68.2 bits (165), Expect = 5e-12
Identities = 53/190 (27%), Positives = 86/190 (44%), Gaps = 2/190 (1%)
Frame = -1
Query: 46 KLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKP-D 104
K+K V +++F M+ G+ P + +NSLI S I A+ L + E +N +P +
Sbjct: 632 KIKMVDEALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVD--EMHDNGQPAN 459
Query: 105 FHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFE 164
TYN I A K+ +VD +A K G+ PD+ TF +I G + A VF+
Sbjct: 458 IFTYNCLIDALCKNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQ 279
Query: 165 EMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNR-WQISETMAAMLVVLYHEQ 223
++ N M+ G C + A + DN + T ++ L+H+
Sbjct: 278 DLLSKGYSVNAWTYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTYETLIQALFHKD 99
Query: 224 GQVEKMEELL 233
+ EK E+LL
Sbjct: 98 -ENEKAEKLL 72
Score = 45.8 bits (107), Expect = 3e-05
Identities = 33/124 (26%), Positives = 55/124 (43%)
Frame = -1
Query: 43 LCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYK 102
LC K +V I + ++ G++P FN LI + A +F+ + S+ Y
Sbjct: 428 LC-KNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQDL-LSKGYS 255
Query: 103 PDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRV 162
+ TYN ++ K G D A S G+ PD T+E++I + E A+++
Sbjct: 254 VNAWTYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTYETLIQALFHKDENEKAEKL 75
Query: 163 FEEM 166
EM
Sbjct: 74 LREM 63
>TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [imported] -
Arabidopsis thaliana, partial (53%)
Length = 1241
Score = 67.8 bits (164), Expect = 6e-12
Identities = 41/170 (24%), Positives = 81/170 (47%)
Frame = +1
Query: 30 NVKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAY 89
++ + G + LI L GK + +F M + + L N+L++A L S
Sbjct: 250 DISNEGFSARLITLYGKSNMHRHAQKLFDEMPQRNCERSVLSLNALLAAYLHSKQYDVVE 429
Query: 90 SLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISG 149
LF+ + + KPD +YN +I A + G+ D+ ++ + G+ DL TF +++ G
Sbjct: 430 RLFKKLPVQLSVKPDLVSYNTYIKALLEKGSFDSAVSVLEEMEKDGVESDLITFNTLLDG 609
Query: 150 CVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFK 199
+ +E ++++E++ ++PN+ + L G K A EF++
Sbjct: 610 LYSKGRFEDGEKLWEKLGEKNVVPNIRTYNARLLGLAVAKRAGEAVEFYE 759
Score = 45.4 bits (106), Expect = 3e-05
Identities = 28/109 (25%), Positives = 49/109 (44%)
Frame = +1
Query: 54 INVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFIS 113
++V ME GV+ + FN+L+ S L+E + +N P+ TYN +
Sbjct: 535 VSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDGEKLWEKL-GEKNVVPNIRTYNARLL 711
Query: 114 AFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRV 162
+ + + +Y + G+ PDL +F ++I G N N + A V
Sbjct: 712 GLAVAKRAGEAVEFYEEMEKKGVKPDLFSFNALIKGFANEGNLDEAKNV 858
>TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Arabidopsis
thaliana}, partial (7%)
Length = 1580
Score = 66.2 bits (160), Expect = 2e-11
Identities = 48/188 (25%), Positives = 81/188 (42%)
Frame = +1
Query: 46 KLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDF 105
K+K +N+F M + P + ++SLI S I A L + M P+
Sbjct: 697 KIKKFDEAMNLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHD-RGVPPNI 873
Query: 106 HTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEE 165
TYN+ + A K+ VD +A + K G PD+ T+ +I G S E A +VFE
Sbjct: 874 CTYNSILDALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEG 1053
Query: 166 MKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQ 225
+ + NV M++GFC + A + DN ++++ ++ +
Sbjct: 1054LLVKGHNLNVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIIILSLFKKDE 1233
Query: 226 VEKMEELL 233
+ E+LL
Sbjct: 1234NDMAEKLL 1257
Score = 57.4 bits (137), Expect = 8e-09
Identities = 38/171 (22%), Positives = 73/171 (42%)
Frame = +1
Query: 64 GVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDA 123
G+KP + +NSL+ ++ A S+F M PD +Y+ I+ F K D
Sbjct: 541 GIKPNFVTYNSLMDGYCLVKEVNKAKSIFNTMAQG-GVNPDIQSYSIMINGFCKIKKFDE 717
Query: 124 MLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLK 183
+ + + PD+ T+ S+I G S A ++ ++M + PN+ S+L
Sbjct: 718 AMNLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILD 897
Query: 184 GFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELLE 234
C + +A D +Q + ++L+ + G++E ++ E
Sbjct: 898 ALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFE 1050
Score = 49.7 bits (117), Expect = 2e-06
Identities = 23/87 (26%), Positives = 47/87 (53%)
Frame = +1
Query: 98 SENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYE 157
++ KP+F TYN+ + + V+ + ++ G+ PD+Q++ +I+G K ++
Sbjct: 535 NKGIKPNFVTYNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFD 714
Query: 158 IADRVFEEMKISEMIPNVTILESMLKG 184
A +F+EM +IP+V S++ G
Sbjct: 715 EAMNLFKEMHRKNIIPDVVTYSSLIDG 795
Score = 38.5 bits (88), Expect = 0.004
Identities = 26/111 (23%), Positives = 51/111 (45%)
Frame = +2
Query: 33 DHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLF 92
D + LI K+ + +++ ++G V+P +++N++I + + A+ LF
Sbjct: 131 DQISYGTLIHGLCKVGETRAALDLLQRVDGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLF 310
Query: 93 EIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTF 143
M S + PD TY+ IS F G + + ++ + PD+ TF
Sbjct: 311 SEMVS-KGISPDVVTYSALISGFCILGKLKDAIDLFNKMILENIKPDVYTF 460
Score = 34.7 bits (78), Expect = 0.058
Identities = 21/75 (28%), Positives = 39/75 (52%), Gaps = 1/75 (1%)
Frame = +2
Query: 102 KPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISG-CVNSKNYEIAD 160
+P+ YN I + K V+ +S + G+ PD+ T+ ++ISG C+ K + D
Sbjct: 230 QPNVVMYNTIIDSMCKVKLVNEAFDLFSEMVSKGISPDVVTYSALISGFCILGKLKDAID 409
Query: 161 RVFEEMKISEMIPNV 175
+F +M + + P+V
Sbjct: 410 -LFNKMILENIKPDV 451
>TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein AT4g19900
[imported] - Arabidopsis thaliana, partial (18%)
Length = 756
Score = 62.4 bits (150), Expect = 3e-10
Identities = 44/187 (23%), Positives = 86/187 (45%), Gaps = 2/187 (1%)
Frame = +1
Query: 20 EWILEKLLKEN-VKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISA 78
E +L ++ ++ V + ++ LI K N + ++ M G P +N++++
Sbjct: 52 EMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMSSEGFSPNLCTYNAIVNG 231
Query: 79 CLSSHDIVTAYSLFEIMESSEN-YKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLG 137
+ AY + E + +N KPD TYN +S K N+ LA ++ G+
Sbjct: 232 LCKRGRVQEAYKMLE--DGFQNGLKPDKFTYNILMSEHCKQENIRQALALFNKMLKIGIQ 405
Query: 138 PDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEF 197
PD+ ++ ++I+ + ++ FEE +IP SM+ G+C + +L A +F
Sbjct: 406 PDIHSYTTLIAVFCRENRMKESEMFFEEAVRIGIIPTNKTYTSMICGYCREGNLTLAMKF 585
Query: 198 FKFVLDN 204
F + D+
Sbjct: 586 FHRLSDH 606
Score = 51.6 bits (122), Expect = 5e-07
Identities = 31/140 (22%), Positives = 57/140 (40%)
Frame = +1
Query: 64 GVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDA 123
G+KP +N L+S +I A +LF M +PD H+Y I+ F + +
Sbjct: 292 GLKPDKFTYNILMSEHCKQENIRQALALFNKMLKI-GIQPDIHSYTTLIAVFCRENRMKE 468
Query: 124 MLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLK 183
++ G+ P +T+ S+I G N +A + F + P +++
Sbjct: 469 SEMFFEEAVRIGIIPTNKTYTSMICGYCREGNLTLAMKFFHRLSDHGCAPESITYGAIIS 648
Query: 184 GFCSQKSLCRANEFFKFVLD 203
G C Q A + +++
Sbjct: 649 GLCKQSKRDEARSLYDSMIE 708
Score = 40.8 bits (94), Expect = 8e-04
Identities = 25/128 (19%), Positives = 56/128 (43%)
Frame = +1
Query: 46 KLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDF 105
K +N++ + +F M +G++P + +LI+ + + + FE P
Sbjct: 343 KQENIRQALALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRI-GIIPTN 519
Query: 106 HTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEE 165
TY + I + + GN+ + ++ G P+ T+ ++ISG + A +++
Sbjct: 520 KTYTSMICGYCREGNLTLAMKFFHRLSDHGCAPESITYGAIISGLCKQSKRDEARSLYDS 699
Query: 166 MKISEMIP 173
M ++P
Sbjct: 700 MIEKGLVP 723
Score = 40.0 bits (92), Expect = 0.001
Identities = 37/191 (19%), Positives = 78/191 (40%), Gaps = 2/191 (1%)
Frame = +1
Query: 108 YNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMK 167
Y I + + ++ S K GL P+ T+ ++I G + N+E A + M
Sbjct: 1 YTAMIRGYCREDKLNRAEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMS 180
Query: 168 ISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVE 227
PN+ +++ G C + R E +K + D + +L E + E
Sbjct: 181 SEGFSPNLCTYNAIVNGLCKRG---RVQEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQE 351
Query: 228 KMEELLETITSYPIDSGVLSQIH--CGIVTMYAMLDRLDEVELSVGRMLKQGMSFTSSDD 285
+ + L + + + G+ IH ++ ++ +R+ E E+ ++ G+ ++
Sbjct: 352 NIRQAL-ALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRIGI-IPTNKT 525
Query: 286 VEKVICSYFRK 296
+IC Y R+
Sbjct: 526 YTSMICGYCRE 558
>TC82835 weakly similar to GP|21741712|emb|CAD41335. oj991113_30.17 {Oryza
sativa}, partial (5%)
Length = 915
Score = 60.5 bits (145), Expect = 1e-09
Identities = 38/150 (25%), Positives = 75/150 (49%), Gaps = 2/150 (1%)
Frame = +3
Query: 22 ILEKLLKENVK--DHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISAC 79
I E L ++N D ++ ++ + G+ V +++F S++ G P ++ +N+LI+
Sbjct: 72 IFEHLREQNNSCLDLITYNIVLDILGRKGRVDEMLDMFASLKETGFVPDTISYNTLINGL 251
Query: 80 LSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPD 139
+ F+ M+ + N +PD TY I ++GN++ L ++ K G+ P
Sbjct: 252 RKVGRSDMCFEYFKEMKENGN-EPDLLTYTALIDISGRAGNIEESLKFFMEMKLKGILPS 428
Query: 140 LQTFESVISGCVNSKNYEIADRVFEEMKIS 169
+Q + S+I ++N E+A + EEM S
Sbjct: 429 IQIYRSLIHNLNKTENIELATELLEEMNSS 518
Score = 50.4 bits (119), Expect = 1e-06
Identities = 42/173 (24%), Positives = 74/173 (42%), Gaps = 3/173 (1%)
Frame = +3
Query: 69 SLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWY 128
S N +I A S +A ++FE + N D TYN + + G VD ML +
Sbjct: 6 SSFINKIIFAFAKSGQKDSALAIFEHLREQNNSCLDLITYNIVLDILGRKGRVDEMLDMF 185
Query: 129 SAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQ 188
++ K TG PD ++ ++I+G ++ F+EMK + P++ +++
Sbjct: 186 ASLKETGFVPDTISYNTLINGLRKVGRSDMCFEYFKEMKENGNEPDLLTYTALID---IS 356
Query: 189 KSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVEKME---ELLETITS 238
E KF ++ + + + L H + E +E ELLE + S
Sbjct: 357 GRAGNIEESLKFFMEMKLKGILPSIQIYRSLIHNLNKTENIELATELLEEMNS 515
>TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16.90 -
Arabidopsis thaliana, partial (43%)
Length = 1038
Score = 58.5 bits (140), Expect = 4e-09
Identities = 45/197 (22%), Positives = 88/197 (43%)
Frame = +1
Query: 37 FSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIME 96
++ LI GK + + + VF M GV+PT +N L+ A S + A +F+ M
Sbjct: 109 YALLINAYGKARREEEALAVFEEMLDAGVRPTRKAYNILLDAFSISGMVEQARIVFKSMR 288
Query: 97 SSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNY 156
+ Y PD +Y +SA+ + +++ ++ G P++ T+ ++I G + +
Sbjct: 289 R-DKYMPDLCSYTTMLSAYVNAPDMEGAEKFFKRLIQDGFEPNVVTYGTLIKGYAKANDI 465
Query: 157 EIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAML 216
E +EEM + N TIL +++ A +FK + N + +L
Sbjct: 466 EKVMEKYEEMLGRGIKANQTILTTIMDAHGKNGDFDSAVNWFKEMALNGLLPDQKAKNIL 645
Query: 217 VVLYHEQGQVEKMEELL 233
+ L + +++ EL+
Sbjct: 646 LSLAKTEEDIKEANELV 696
Score = 57.4 bits (137), Expect = 8e-09
Identities = 37/173 (21%), Positives = 84/173 (48%)
Frame = +1
Query: 55 NVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISA 114
N++ M+ ++P + + LI+A + A ++FE M + +P YN + A
Sbjct: 58 NIYDQMQRADLRPDVVSYALLINAYGKARREEEALAVFEEMLDA-GVRPTRKAYNILLDA 234
Query: 115 FSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPN 174
FS SG V+ + + + PDL ++ +++S VN+ + E A++ F+ + PN
Sbjct: 235 FSISGMVEQARIVFKSMRRDKYMPDLCSYTTMLSAYVNAPDMEGAEKFFKRLIQDGFEPN 414
Query: 175 VTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVE 227
V +++KG+ + + E ++ +L + ++T+ ++ + + G +
Sbjct: 415 VVTYGTLIKGYAKANDIEKVMEKYEEMLGRGIKANQTILTTIMDAHGKNGDFD 573
Score = 47.8 bits (112), Expect = 7e-06
Identities = 34/166 (20%), Positives = 76/166 (45%)
Frame = +1
Query: 69 SLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWY 128
++ +NSL+S + ++ Y + + +PD +Y I+A+ K+ + LA +
Sbjct: 4 TVTYNSLMSFETNYKEVSNIYDQMQ----RADLRPDVVSYALLINAYGKARREEEALAVF 171
Query: 129 SAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQ 188
G+ P + + ++ S E A VF+ M+ + +P++ +ML + +
Sbjct: 172 EEMLDAGVRPTRKAYNILLDAFSISGMVEQARIVFKSMRRDKYMPDLCSYTTMLSAYVNA 351
Query: 189 KSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELLE 234
+ A +FFK ++ + ++ + L+ Y + +EK+ E E
Sbjct: 352 PDMEGAEKFFKRLIQDGFEPNVVTYGTLIKGYAKANDIEKVMEKYE 489
Score = 30.8 bits (68), Expect = 0.83
Identities = 33/172 (19%), Positives = 72/172 (41%)
Frame = +1
Query: 107 TYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEM 166
TYN+ +S + V + Y + L PD+ ++ +I+ ++ E A VFEEM
Sbjct: 10 TYNSLMSFETNYKEVSNI---YDQMQRADLRPDVVSYALLINAYGKARREEEALAVFEEM 180
Query: 167 KISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQV 226
+ + P +L F + +A FK + +++ ++ Y +
Sbjct: 181 LDAGVRPTRKAYNILLDAFSISGMVEQARIVFKSMRRDKYMPDLCSYTTMLSAYVNAPDM 360
Query: 227 EKMEELLETITSYPIDSGVLSQIHCGIVTMYAMLDRLDEVELSVGRMLKQGM 278
E E+ + + + V++ + ++ YA + +++V ML +G+
Sbjct: 361 EGAEKFFKRLIQDGFEPNVVT--YGTLIKGYAKANDIEKVMEKYEEMLGRGI 510
>BF649699 similar to GP|9757911|dbj gb|AAF19552.1~gene_id:MMN10.14~similar to
unknown protein {Arabidopsis thaliana}, partial (11%)
Length = 617
Score = 56.6 bits (135), Expect = 1e-08
Identities = 43/194 (22%), Positives = 78/194 (40%), Gaps = 34/194 (17%)
Frame = +1
Query: 46 KLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMES-------- 97
K N+ ++ + G P V+N+LI+A D+ A L++ M S
Sbjct: 10 KKGNIDSAYDLVVKLGRFGFLPNLFVYNALINALCKGEDLDKAELLYKNMHSMNLPLNDV 189
Query: 98 -----------------SENY---------KPDFHTYNNFISAFSKSGNVDAMLAWYSAK 131
+E+Y + + YN+ I+ K G++ A Y+
Sbjct: 190 TYSILIDSFCKRGMLDVAESYFGRMIEDGIRETIYPYNSLINGHCKFGDLSAAEFLYTKM 369
Query: 132 KATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSL 191
GL P TF ++ISG E A +++ EM E P+V +++ G CS +
Sbjct: 370 INEGLEPTATTFTTLISGYCKDLQVEKAFKLYREMNEKEXAPSVYTFTALIYGLCSTNEM 549
Query: 192 CRANEFFKFVLDNR 205
A++ F +++ +
Sbjct: 550 AEASKLFDEMVERK 591
>BG586292 similar to PIR|T46163|T46 nodulin / glutamate-ammonia ligase-like
protein - Arabidopsis thaliana, partial (50%)
Length = 719
Score = 55.5 bits (132), Expect = 3e-08
Identities = 35/149 (23%), Positives = 66/149 (43%)
Frame = -3
Query: 37 FSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIME 96
F+ LI C KL L V M +GV+ +++ +N+++ + ++ M
Sbjct: 504 FTVLISCCCKLARFDLVNRVLLEMSYLGVRCSTVTYNTILDGYGKAGLFEEMENVLADMI 325
Query: 97 SSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNY 156
+ PD T N+ I ++ GN+ M +WY+ + G+ PD+ TF +I + Y
Sbjct: 324 EDGDSLPDVFTLNSIIGSYGNGGNIRKMESWYNRFQLMGVEPDITTFNVLILSFGKAGMY 145
Query: 157 EIADRVFEEMKISEMIPNVTILESMLKGF 185
+ V + M+ P V +++ F
Sbjct: 144 KKMSSVMDYMEKRFFSPTVVTYNIVIETF 58
Score = 44.3 bits (103), Expect = 7e-05
Identities = 37/175 (21%), Positives = 79/175 (45%), Gaps = 6/175 (3%)
Frame = -3
Query: 66 KPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAML 125
KPT V+ SLIS + A++ E M+S+ + PD T+ IS K D +
Sbjct: 630 KPTIDVYTSLISVYGKCSLLEKAFATLEYMKSASDCTPDVFTFTVLISCCCKLARFDLVN 451
Query: 126 AWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEM-KISEMIPNVTILESMLKG 184
G+ T+ +++ G + +E + V +M + + +P+V L S++
Sbjct: 450 RVLLEMSYLGVRCSTVTYNTILDGYGKAGLFEEMENVLADMIEDGDSLPDVFTLNSIIGS 271
Query: 185 FCSQKSLCRANEFFKFVLDNRWQI-----SETMAAMLVVLYHEQGQVEKMEELLE 234
+ + ++ + ++ NR+Q+ T +L++ + + G +KM +++
Sbjct: 270 YGNGGNIRKMESWY-----NRFQLMGVEPDITTFNVLILSFGKAGMYKKMSSVMD 121
>BE998743 similar to GP|22128591|gb| fertility restorer-like protein {Petunia
x hybrida}, partial (7%)
Length = 829
Score = 54.3 bits (129), Expect = 7e-08
Identities = 40/185 (21%), Positives = 81/185 (43%), Gaps = 1/185 (0%)
Frame = +2
Query: 55 NVFTSMEGVGVKPTSLVFNSLISA-CLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFIS 113
+V +++ +GV P + +N +++ C SH A+ L + M + PD TY++ I
Sbjct: 239 HVLSTIARMGVAPDAQSYNIMVNGFCRISH----AWKLVDEMHVN-GQPPDIFTYSSLID 403
Query: 114 AFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIP 173
A K+ ++D +A K G+ P++ T+ +I G + A VF+++
Sbjct: 404 ALCKNNHLDKAIALVKKIKDQGIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTKGYSL 583
Query: 174 NVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELL 233
N+ ++ G C + +A + DN + ++ + EK E+LL
Sbjct: 584 NIRTYNILINGLCKEGLFDKAEALLSKMEDNDINPNVVTYETIIRSLFYKDYNEKAEKLL 763
Query: 234 ETITS 238
+ +
Sbjct: 764 REMVA 778
Score = 50.1 bits (118), Expect = 1e-06
Identities = 41/201 (20%), Positives = 87/201 (42%), Gaps = 2/201 (0%)
Frame = +2
Query: 49 NVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTY 108
NV+ N M +KP + +NSL+ +++ A + + + PD +Y
Sbjct: 116 NVKGAKNALAMMIKGSIKPDVVTYNSLMDGYCLVNEVNKAKHVLSTI-ARMGVAPDAQSY 292
Query: 109 NNFISAFSKSGNVDAMLAWYSAKK--ATGLGPDLQTFESVISGCVNSKNYEIADRVFEEM 166
N ++ F + + AW + G PD+ T+ S+I + + + A + +++
Sbjct: 293 NIMVNGFCRISH-----AWKLVDEMHVNGQPPDIFTYSSLIDALCKNNHLDKAIALVKKI 457
Query: 167 KISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQV 226
K + PN+ ++ G C L A + F+ +L + ++ +L+ ++G
Sbjct: 458 KDQGIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTKGYSLNIRTYNILINGLCKEGLF 637
Query: 227 EKMEELLETITSYPIDSGVLS 247
+K E LL + I+ V++
Sbjct: 638 DKAEALLSKMEDNDINPNVVT 700
>TC83856 weakly similar to PIR|B96656|B96656 unknown protein 41955-40111
[imported] - Arabidopsis thaliana, partial (12%)
Length = 662
Score = 53.5 bits (127), Expect = 1e-07
Identities = 34/124 (27%), Positives = 54/124 (43%)
Frame = +2
Query: 65 VKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAM 124
+KP FN L+ S I A L + M P+ TY++ + A K+ VD
Sbjct: 68 IKPDVYTFNILVDVFCKSGKISYALKLVDEMHD-RGQPPNIVTYSSILDALCKTHRVDKA 244
Query: 125 LAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKG 184
+A + K G+ P++ T+ +I G S E A +FE++ + V M G
Sbjct: 245 VALLTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDITVVTYIVMFYG 424
Query: 185 FCSQ 188
FC +
Sbjct: 425 FCKK 436
Score = 52.0 bits (123), Expect = 3e-07
Identities = 30/122 (24%), Positives = 55/122 (44%)
Frame = +2
Query: 99 ENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEI 158
EN KPD +T+N + F KSG + L G P++ T+ S++ + +
Sbjct: 62 ENIKPDVYTFNILVDVFCKSGKISYALKLVDEMHDRGQPPNIVTYSSILDALCKTHRVDK 241
Query: 159 ADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVV 218
A + ++K + PN+ ++ G C+ L A F+ +L + I T+ +V+
Sbjct: 242 AVALLTKLKDQGIRPNMHTYTILIDGLCTSGKLEDARNIFEDLLVKGYDI--TVVTYIVM 415
Query: 219 LY 220
Y
Sbjct: 416 FY 421
>BF635590 weakly similar to PIR|D84778|D84 probable salt-inducible protein
[imported] - Arabidopsis thaliana, partial (25%)
Length = 465
Score = 52.8 bits (125), Expect = 2e-07
Identities = 38/138 (27%), Positives = 62/138 (44%), Gaps = 3/138 (2%)
Frame = +3
Query: 92 FEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCV 151
F +M + KP N I F KSG D +Y+ + PD+ TF +ISG
Sbjct: 60 FNVMRRLIDGKPSVSVCNILIHGFVKSGRFDRAFEFYNQMVKDRIKPDVFTFNILISGYC 239
Query: 152 NSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFV---LDNRWQI 208
+ A +F+EM+ PNV +++KG + CR +E V ++ Q+
Sbjct: 240 RDFKFGFALEMFDEMRKMGCHPNVVTFNTLIKGLFRE---CRVDEGIGMVYEMIELGCQL 410
Query: 209 SETMAAMLVVLYHEQGQV 226
S+ +LV ++G+V
Sbjct: 411 SDVTCEILVDGLCKEGRV 464
>TC82633 similar to GP|15450347|gb|AAK96467.1 At1g20300/F14O10_8
{Arabidopsis thaliana}, partial (34%)
Length = 830
Score = 52.4 bits (124), Expect = 3e-07
Identities = 40/190 (21%), Positives = 87/190 (45%), Gaps = 5/190 (2%)
Frame = +1
Query: 54 INVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFIS 113
+ V+ M+ +G ++ +N LI + ++ A + + M + P+ T+N+
Sbjct: 10 LQVYNQMKRLGCAADTISYNFLIESHCKDENLDEAVKVLDTMVK-KGVAPNASTFNSIFG 186
Query: 114 AFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIP 173
++ +V+ Y+ K P+ T+ ++ +SK+ ++ ++ +EM SE+ P
Sbjct: 187 CIAELHDVNGAHRMYAKMKELKCMPNTLTYNILMRMFADSKSIDMVLKLKKEMDESEVEP 366
Query: 174 NVTILESMLKGFCSQKSLCRANEFFKFVLDNR-----WQISETMAAMLVVLYHEQGQVEK 228
NV ++ FC + A K +++ + I ET+ +L GQ++K
Sbjct: 367 NVNTYRILILMFCEKGHWNNAYNLMKEMVEEKCLKPNLSIYETVLELL----RNAGQLKK 534
Query: 229 MEELLETITS 238
EEL+E + +
Sbjct: 535 HEELVEKMVA 564
Score = 48.9 bits (115), Expect = 3e-06
Identities = 33/139 (23%), Positives = 68/139 (48%), Gaps = 1/139 (0%)
Frame = +1
Query: 46 KLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDF 105
K +N+ + V +M GV P + FNS+ HD+ A+ ++ M+ + P+
Sbjct: 91 KDENLDEAVKVLDTMVKKGVAPNASTFNSIFGCIAELHDVNGAHRMYAKMKELK-CMPNT 267
Query: 106 HTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEE 165
TYN + F+ S ++D +L + + P++ T+ +I ++ A + +E
Sbjct: 268 LTYNILMRMFADSKSIDMVLKLKKEMDESEVEPNVNTYRILILMFCEKGHWNNAYNLMKE 447
Query: 166 MKISEMI-PNVTILESMLK 183
M + + PN++I E++L+
Sbjct: 448 MVEEKCLKPNLSIYETVLE 504
>TC80520 weakly similar to PIR|T02562|T02562 probable salt-inducible protein
[imported] - Arabidopsis thaliana, partial (6%)
Length = 1911
Score = 52.0 bits (123), Expect = 3e-07
Identities = 44/192 (22%), Positives = 77/192 (39%), Gaps = 36/192 (18%)
Frame = +1
Query: 43 LCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIM-ESSENY 101
L G+ V+ +VF S++ VG P++ FN + ACL + L+E+M ES
Sbjct: 391 LLGENGMVEEVFDVFVSLKKVGFLPSASSFNVCLLACLKVGRTDLVWKLYELMIESGVGV 570
Query: 102 KPDFHTYNNFISAFSKSGNV----------------------DAMLAWYSAKK------- 132
D T I AF V +A++ + +K
Sbjct: 571 NIDVETVGCLIKAFCAENKVFNGYELLRQVLEKGLCVDNTVFNALINGFCKQKQYDRVSE 750
Query: 133 ------ATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFC 186
A P + T++ +I+G + + + A RVF ++K P+ + +++KGFC
Sbjct: 751 ILHIMIAMKCNPSIYTYQEIINGLLKRRKNDEAFRVFNDLKDRGYFPDRVMYTTVIKGFC 930
Query: 187 SQKSLCRANEFF 198
L A + +
Sbjct: 931 DMGLLAEARKLW 966
Score = 29.3 bits (64), Expect = 2.4
Identities = 21/100 (21%), Positives = 42/100 (42%)
Frame = +3
Query: 135 GLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRA 194
G ++ ++ ++ISG + A +F EM + ++ S++KG C + L +A
Sbjct: 1089 GYAENVVSYSTMISGLYLHGKTDEALSLFHEMSRKGIARDLISYNSLIKGLCQEGELAKA 1268
Query: 195 NEFFKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELLE 234
+L + S + L+ + G E LL+
Sbjct: 1269 TNLLNKLLIQGLEPSVSSFTPLIKCLCKVGDTEGAMRLLK 1388
>BI270314 similar to GP|9759030|dbj contains similarity to salt-inducible
protein~gene_id:K6A12.14 {Arabidopsis thaliana}, partial
(30%)
Length = 669
Score = 51.6 bits (122), Expect = 5e-07
Identities = 44/171 (25%), Positives = 75/171 (43%), Gaps = 7/171 (4%)
Frame = +3
Query: 22 ILEKLLKENVKDHG------RFSELIFLCGKLKNVQ-LGINVFTSMEGVGVKPTSLVFNS 74
I+E LL E +KD G ++ LI G+ K + + + F M+ VG+KPTS + +
Sbjct: 66 IVESLLAE-MKDFGLKPNANSYTCLISAYGRQKKMSDMAADAFLKMKKVGIKPTSHSYTA 242
Query: 75 LISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKAT 134
+I A S AY++FE M E KP TY + AF + G+ + ++ + +
Sbjct: 243 MIHAYSVSGWHEKAYAVFENM-IREGIKPSIETYTTLLDAFRRVGDTETLMKIWKLMMSE 419
Query: 135 GLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGF 185
+ TF ++ G + A V E + P V ++ +
Sbjct: 420 KVKGTQVTFNILVDGFAKQGLFMEARDVISEFGKIGLQPTVMTYNMLINAY 572
Score = 37.7 bits (86), Expect = 0.007
Identities = 36/198 (18%), Positives = 77/198 (38%)
Frame = +3
Query: 37 FSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIME 96
F+ L++ + ++ ++ M+ G+KP + + LISA + + +
Sbjct: 21 FNILMYAYSRRMQPKIVESLLAEMKDFGLKPNANSYTCLISAYGRQKKMSDMAADAFLKM 200
Query: 97 SSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNY 156
KP H+Y I A+S SG + A + G+ P ++T+ +++ +
Sbjct: 201 KKVGIKPTSHSYTAMIHAYSVSGWHEKAYAVFENMIREGIKPSIETYTTLLDAFRRVGDT 380
Query: 157 EIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAML 216
E ++++ M ++ ++ GF Q A + Q + ML
Sbjct: 381 ETLMKIWKLMMSEKVKGTQVTFNILVDGFAKQGLFMEARDVISEFGKIGLQPTVMTYNML 560
Query: 217 VVLYHEQGQVEKMEELLE 234
+ Y G + +LL+
Sbjct: 561 INAYARGGLDSNIPQLLK 614
>TC83956 weakly similar to GP|15982931|gb|AAL09812.1 AT3g53700/F4P12_400
{Arabidopsis thaliana}, partial (18%)
Length = 762
Score = 51.6 bits (122), Expect = 5e-07
Identities = 41/200 (20%), Positives = 91/200 (45%), Gaps = 2/200 (1%)
Frame = +1
Query: 60 MEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSG 119
++ G P + F +LI + + H+I +L +I+E+ +KPD + YN ++A +
Sbjct: 166 LKSSGSIPNATTFATLIQSFTNFHEIE---NLLKILENELGFKPDTNFYNIALNALVEDN 336
Query: 120 NVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILE 179
+ + +S G+ D+ TF +I + A + EEM + P+
Sbjct: 337 KLKLVEMLHSKMVNEGIVLDVSTFNVLIKALCKAHQLRPAILMLEEMANHGLKPDEITFT 516
Query: 180 SMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELLETIT-- 237
++++GF + L A + K +L ++ +LV + ++G+VE+ + ++
Sbjct: 517 TLMQGFIEEGDLNGALKMKKQMLGYGCLLTNVSVKVLVNGFCKEGRVEEALRFVLEVSEE 696
Query: 238 SYPIDSGVLSQIHCGIVTMY 257
+ D G + + G+ ++
Sbjct: 697 GFSPDQGTFNSLVNGVCRIW 756
>TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Arabidopsis
thaliana}, partial (7%)
Length = 978
Score = 50.4 bits (119), Expect = 1e-06
Identities = 44/223 (19%), Positives = 94/223 (41%), Gaps = 3/223 (1%)
Frame = +1
Query: 48 KNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHT 107
+N + V+ SM G+K ++F+ ++ + +FE + S + D
Sbjct: 109 RNFDKALAVYKSMISRGIKTNCVIFSCILHCLDEMGRALEVVDMFEEFKESGLF-IDRKA 285
Query: 108 YNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMK 167
YN A K G VD + K+ L D++ + ++I+G A +F+EM+
Sbjct: 286 YNILFDALCKLGKVDDAVGMLDELKSMQLDVDMKHYTTLINGYFLQGKPIEAQSLFKEME 465
Query: 168 ISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVE 227
P+V + GF ++ A + ++ + + T +++ G+VE
Sbjct: 466 ERGFKPDVVAYNVLAAGFFRNRTDFEAMDLLNYMESQGVEPNSTTHKIIIEGLCSAGKVE 645
Query: 228 KMEELLETITSYPIDSGVLSQIHCGIVTMY---AMLDRLDEVE 267
+ EE + ++ V +I+ +V Y A++++ E++
Sbjct: 646 EAEEFFNWLKGESVEISV--EIYTALVNGYCEAALIEKSHELK 768
>BF633747 similar to GP|10177016|db maize crp1 protein-like {Arabidopsis
thaliana}, partial (21%)
Length = 537
Score = 49.3 bits (116), Expect = 2e-06
Identities = 30/115 (26%), Positives = 55/115 (47%)
Frame = +1
Query: 60 MEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSG 119
++ +G KPTS ++N+LI+A A + F +M ++E P N+ I+AF +
Sbjct: 37 LKSMGFKPTSTMYNALINAYAQRGLSDQAVNAFRMM-AAEGLTPSLLALNSLINAFGEDR 213
Query: 120 NVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPN 174
A K G+ PD+ T+ +++ + + V+EEM +S P+
Sbjct: 214 RDAEAFAVLQYMKENGVKPDVVTYTTLMKALIRVDKFHKVPAVYEEMVMSGCTPD 378
>BF004907 weakly similar to GP|8953393|emb putative protein {Arabidopsis
thaliana}, partial (34%)
Length = 665
Score = 49.3 bits (116), Expect = 2e-06
Identities = 37/133 (27%), Positives = 58/133 (42%)
Frame = +3
Query: 35 GRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEI 94
GRF E + + + +Q+G PT +NSL+ + DI A + +
Sbjct: 174 GRFKEALGMMERFHVLQIG-------------PTLSTYNSLVKGFCKAGDIEGASKILKK 314
Query: 95 MESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSK 154
M S + P TYN F FS+ G VD + Y+ +G PD T+ V+ +
Sbjct: 315 M-ISRGFLPIPTTYNYFFRYFSRCGKVDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEE 491
Query: 155 NYEIADRVFEEMK 167
E+A +V EM+
Sbjct: 492 KLELAVQVSMEMR 530
Score = 41.2 bits (95), Expect = 6e-04
Identities = 26/121 (21%), Positives = 51/121 (41%)
Frame = +3
Query: 65 VKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAM 124
V+P+ + + +L+ + A + M + E KP+ YN I A +++G
Sbjct: 15 VRPSVVTYGTLVEGYCRMRRVEKALEMVGEM-TKEGIKPNAIVYNPIIDALAEAGRFKEA 191
Query: 125 LAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKG 184
L +GP L T+ S++ G + + E A ++ ++M +P T +
Sbjct: 192 LGMMERFHVLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFFRY 371
Query: 185 F 185
F
Sbjct: 372 F 374
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,789,085
Number of Sequences: 36976
Number of extensions: 105808
Number of successful extensions: 715
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 624
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 685
length of query: 356
length of database: 9,014,727
effective HSP length: 97
effective length of query: 259
effective length of database: 5,428,055
effective search space: 1405866245
effective search space used: 1405866245
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148532.2


