

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148528.7 - phase: 0 /pseudo
(1391 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
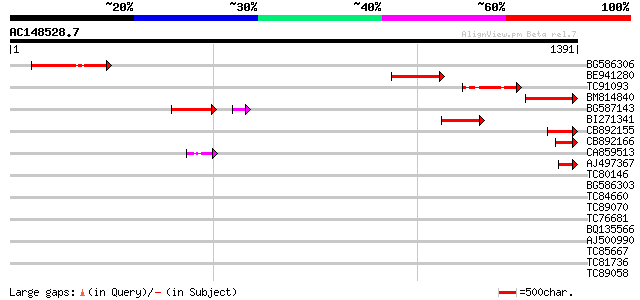
Score E
Sequences producing significant alignments: (bits) Value
BG586306 weakly similar to GP|15128241|db helicase-like protein ... 173 4e-43
BE941280 weakly similar to GP|20197614|g unknown protein {Arabid... 153 4e-37
TC91093 weakly similar to PIR|T01873|T01873 hypothetical protein... 147 3e-35
BM814840 weakly similar to GP|15128241|db helicase-like protein ... 145 8e-35
BG587143 weakly similar to PIR|A96586|A9 hypothetical protein F2... 120 6e-31
BI271341 112 1e-24
CB892155 similar to PIR|D86481|D86 hypothetical protein AAG28292... 90 5e-18
CB892166 similar to GP|20197614|gb unknown protein {Arabidopsis ... 58 3e-08
CA859513 52 1e-06
AJ497367 similar to GP|14140286|gb putative helicase {Oryza sati... 52 1e-06
TC80146 36 0.12
BG586303 weakly similar to PIR|A96586|A96 hypothetical protein F... 35 0.26
TC84660 similar to PIR|D86481|D86481 hypothetical protein AAG282... 33 0.59
TC89070 similar to GP|20197343|gb|AAM15033.1 putative helicase {... 32 1.3
TC76681 homologue to SP|O22585|AMYB_MEDSA Beta-amylase (EC 3.2.1... 31 2.9
BQ135566 31 3.8
AJ500990 similar to PIR|T51278|T512 hypothetical protein T8M16_2... 30 5.0
TC85667 similar to GP|15809788|gb|AAL06822.1 AT5g39740/MKM21_30 ... 30 5.0
TC81736 similar to SP|O49204|KAPS_CATRO Adenylylsulfate kinase ... 30 5.0
TC89058 similar to GP|19071218|gb|AAL84162.1 putative heavy meta... 30 6.5
>BG586306 weakly similar to GP|15128241|db helicase-like protein {Oryza
sativa (japonica cultivar-group)}, partial (2%)
Length = 667
Score = 173 bits (439), Expect = 4e-43
Identities = 91/201 (45%), Positives = 137/201 (67%), Gaps = 5/201 (2%)
Frame = +2
Query: 54 FNQNIRLYNSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQL 113
F IR+YNS+ AF+S G K+D V+ G TIRIQGQ+ HR+ S+IP G+ P++ Q+
Sbjct: 11 FRDTIRVYNSVLAFTSIGMKMDYSVVNAPGRYTIRIQGQTHHRIDSLIPRQGRPPEYLQI 190
Query: 114 YIFDTDNELQNRV----QGIRNPNLNMQTVSKLQQMLDDTNCHAKSFRKARDRLRQGNVE 169
YIFDT NE++NR+ Q NL+ T+++L +M+D+ NC AK FR+ARD + G+ +
Sbjct: 191 YIFDTGNEVRNRLNAMGQTSTEGNLDETTLARLIEMIDENNCLAKLFRRARDYYK-GSGQ 367
Query: 170 NLKLKLISDRTTDGRIYNQPTVSEVAALIVGDVDSA-ARRDIIMERQSGRLERIDEFHPA 228
++L+SD+ G+ Y+ P+ SEVA LIVGD+ S RDI+++ QS L++I ++H
Sbjct: 368 EFNIRLLSDK-GKGKEYDLPSTSEVAGLIVGDMSSTIGVRDIVVQFQSDTLQQIRDYHSL 544
Query: 229 YLAYQYPLLFPYGEDGYRDDV 249
Y++ QYPLLFPYGE G+ ++
Sbjct: 545 YMSLQYPLLFPYGEYGFHPEI 607
>BE941280 weakly similar to GP|20197614|g unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 403
Score = 153 bits (387), Expect = 4e-37
Identities = 78/134 (58%), Positives = 98/134 (72%), Gaps = 2/134 (1%)
Frame = -1
Query: 936 LHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIASLLLPGGRTAHSKFKIPVPSFENS 995
L+ YGGT KTF+W L+A+LRS+G+IVL A+SGI +LL+PGGRTAHS+F IP E S
Sbjct: 403 LYDYGGTEKTFIWRALSAALRSEGEIVLACASSGIDALLMPGGRTAHSRFGIPFIIDETS 224
Query: 996 TCNIDGDSDLAKLLKVTNLIIWDEAPMAHKFCFEALDRTLKDIMS--DTADGDKIFGGKV 1053
C + + LA L+ LIIWDEAPM HK CFEALDR+L+D++ D + D FGGKV
Sbjct: 223 MCGVTPNIPLASLVIKAKLIIWDEAPMMHKHCFEALDRSLRDVLKTVDERNKDIPFGGKV 44
Query: 1054 IVFGGDFRQILPVV 1067
+V GGDFRQIL V+
Sbjct: 43 VVLGGDFRQILLVM 2
>TC91093 weakly similar to PIR|T01873|T01873 hypothetical protein T24M8.10 -
Arabidopsis thaliana, partial (11%)
Length = 701
Score = 147 bits (371), Expect = 3e-35
Identities = 80/146 (54%), Positives = 103/146 (69%)
Frame = +3
Query: 1111 ELKTFSEWILKVGEGKISEPNDGIVDFEIPDDLLIKEFDDPIEAIMKSTYPNFLNMYNNP 1170
E KTF+E + GK+SEPND + + P DPI+AI++STYPN ++ YNN
Sbjct: 306 EFKTFAEILT----GKMSEPNDSYAEVDTPPG-------DPIDAIVQSTYPNLVSQYNNE 452
Query: 1171 DYLQQRAILASTIDVVDKINDYVLSIIPGEEKEYFSSDSIDRSEVNDQCQSFQLFTPEFL 1230
+LQ RAIL ST +VVD+INDYVL +IPGEE+ +S+ +RSEVND Q+F PEFL
Sbjct: 453 QFLQSRAILTSTDEVVDQINDYVLKLIPGEERVIYSA---NRSEVND-VQAFDAIPPEFL 620
Query: 1231 STLRTSGLPNHKIKLKVGTPIMLLRN 1256
+L+TS LPNHK+ LKVGTPIMLLR+
Sbjct: 621 QSLKTSDLPNHKLTLKVGTPIMLLRD 698
>BM814840 weakly similar to GP|15128241|db helicase-like protein {Oryza sativa
(japonica cultivar-group)}, partial (6%)
Length = 733
Score = 145 bits (367), Expect = 8e-35
Identities = 72/127 (56%), Positives = 96/127 (74%)
Frame = +3
Query: 1265 NGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAM 1324
+GTRLI+ + +VI A++I G + G ++YIPRM++ PS + R QFP+++S+AM
Sbjct: 27 HGTRLIIVSLGKNVICARVIGGTHAGEVSYIPRMNLIPSGANVSITFERCQFPLVLSFAM 206
Query: 1325 TINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVY 1384
TINKSQGQTL SVGLYLPR VF+HGQLYVA SRV++++GLKILI D P S+T NVVY
Sbjct: 207 TINKSQGQTLTSVGLYLPRPVFTHGQLYVAVSRVKSRSGLKILITDENGSPSSSTVNVVY 386
Query: 1385 KEVFDNL 1391
+EVF +
Sbjct: 387 QEVFQKI 407
>BG587143 weakly similar to PIR|A96586|A9 hypothetical protein F20D21.24
[imported] - Arabidopsis thaliana, partial (12%)
Length = 717
Score = 120 bits (302), Expect(2) = 6e-31
Identities = 56/111 (50%), Positives = 75/111 (67%)
Frame = -3
Query: 397 GLKPHDRPDIIARVFKIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHP 456
G P+D+PDI RVFK+K DELL D K ++ IEFQKRGL HAHIL++
Sbjct: 703 GDSPNDKPDIECRVFKMKLDELLKDFKKGTFFKPYTTALHRIEFQKRGLRHAHILLWFGN 524
Query: 457 SSKYPTPDHINQIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCMK 507
SS+ P+ + +++IISAE+P+ + D E Y LV HM+HGPCG+ P SPCM+
Sbjct: 523 SSRTPSSEEVDEIISAELPNKKQDPEAYNLVTKHMIHGPCGVINPKSPCME 371
Score = 33.1 bits (74), Expect(2) = 6e-31
Identities = 18/45 (40%), Positives = 26/45 (57%)
Frame = -2
Query: 546 KWNQLG*QICCATQSIFIDEISSSYKYGMV*SIKFNQILIQIY*Q 590
KW + C + E+ SSY YGMV*S K +++LIQ++ Q
Sbjct: 257 KWGNPKQHVHCTS*H*APKEVRSSY*YGMV*SYKCSEVLIQVHNQ 123
>BI271341
Length = 468
Score = 112 bits (280), Expect = 1e-24
Identities = 58/104 (55%), Positives = 75/104 (71%)
Frame = +1
Query: 1060 FRQILPVVPRADRSDIINSSINSSYIWDECIVLTLTKNMRLRFNVGSSDADELKTFSEWI 1119
FR ILPV+PR RSDII+++INSS I D C V+ L KNM L+ N SS+ E + FS+ I
Sbjct: 1 FR*ILPVIPRGSRSDIIHATINSSCI*DHCQVVRLKKNMWLQQNGQSSNDPEFEQFSK*I 180
Query: 1120 LKVGEGKISEPNDGIVDFEIPDDLLIKEFDDPIEAIMKSTYPNF 1163
LKVG+GKI EPND D +IP +LLI +DD ++ I++STY NF
Sbjct: 181 LKVGDGKIYEPNDSYADIDIPPELLISNYDDSLQTIVQSTYQNF 312
>CB892155 similar to PIR|D86481|D86 hypothetical protein AAG28292.1 [imported]
- Arabidopsis thaliana, partial (4%)
Length = 572
Score = 90.1 bits (222), Expect = 5e-18
Identities = 46/72 (63%), Positives = 57/72 (78%)
Frame = -2
Query: 1320 VSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNT 1379
V +AMTINKSQGQ+L +G+YLP SVFSHGQLYVA SRV ++ GLKILI + + + T
Sbjct: 364 VYFAMTINKSQGQSLKHIGVYLPSSVFSHGQLYVALSRVTSREGLKILISNDDGEDDCVT 185
Query: 1380 TNVVYKEVFDNL 1391
+NVVY+EVF NL
Sbjct: 184 SNVVYREVFHNL 149
>CB892166 similar to GP|20197614|gb unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 748
Score = 57.8 bits (138), Expect = 3e-08
Identities = 31/52 (59%), Positives = 37/52 (70%)
Frame = -1
Query: 1340 YLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVYKEVFDNL 1391
+ R VFSHGQLYVA SRV ++ GLKIL+ D + NTTNVVYK VF N+
Sbjct: 289 FYDREVFSHGQLYVAISRVSSRNGLKILMIDENGDCIDNTTNVVYK-VFQNV 137
>CA859513
Length = 363
Score = 52.4 bits (124), Expect = 1e-06
Identities = 28/76 (36%), Positives = 42/76 (54%)
Frame = +1
Query: 433 AYMYTIEFQKRGLPHAHILIFLHPSSKYPTPDHINQIISAEIPHPENDRELYKLVGTHMM 492
+++Y EF+K LPHAH+L H + Y T +SAE+ P LY+ V + M+
Sbjct: 130 SHIYVAEFEKCDLPHAHMLF--HRADNYDT------FVSAELSDPVEQLRLYQTVVSVMI 285
Query: 493 HGPCGLAKPSSPCMKK 508
HGP G + PCM++
Sbjct: 286 HGPYGPFNNNVPCMRE 333
>AJ497367 similar to GP|14140286|gb putative helicase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 543
Score = 52.4 bits (124), Expect = 1e-06
Identities = 26/46 (56%), Positives = 35/46 (75%)
Frame = +1
Query: 1346 FSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVYKEVFDNL 1391
FS+G+LYVA SRV ++ GLKIL+ + ++ T+NVVYKEVF NL
Sbjct: 19 FSNGKLYVAVSRVTSRKGLKILLAHEDGNCMNTTSNVVYKEVFRNL 156
>TC80146
Length = 476
Score = 35.8 bits (81), Expect = 0.12
Identities = 16/35 (45%), Positives = 24/35 (67%)
Frame = -3
Query: 1318 IIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLY 1352
I + Y TINKS+ Q+L + +YL R +FSH ++Y
Sbjct: 471 IQIPYFKTINKSR*QSLSYMKIYLSRPIFSHEEMY 367
>BG586303 weakly similar to PIR|A96586|A96 hypothetical protein F20D21.24
[imported] - Arabidopsis thaliana, partial (3%)
Length = 698
Score = 34.7 bits (78), Expect = 0.26
Identities = 14/25 (56%), Positives = 20/25 (80%)
Frame = -3
Query: 400 PHDRPDIIARVFKIKFDELLNDLTK 424
P+DR D+ +RVFK+K DEL++D K
Sbjct: 669 PNDRHDLKSRVFKMKLDELMSDFNK 595
>TC84660 similar to PIR|D86481|D86481 hypothetical protein AAG28292.1
[imported] - Arabidopsis thaliana, partial (1%)
Length = 1009
Score = 33.5 bits (75), Expect = 0.59
Identities = 14/24 (58%), Positives = 19/24 (78%)
Frame = +2
Query: 1338 GLYLPRSVFSHGQLYVAFSRVRTK 1361
G+YLP+ +F HG LYVA SRV ++
Sbjct: 83 GMYLPQPIF*HG*LYVALSRVTSR 154
>TC89070 similar to GP|20197343|gb|AAM15033.1 putative helicase {Arabidopsis
thaliana}, partial (38%)
Length = 861
Score = 32.3 bits (72), Expect = 1.3
Identities = 21/57 (36%), Positives = 27/57 (46%)
Frame = +2
Query: 915 QRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIA 971
Q+D K +S+ N VF LHG GTGKT + +G +L A S IA
Sbjct: 632 QKDAISKALSSKN-----VFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIA 787
>TC76681 homologue to SP|O22585|AMYB_MEDSA Beta-amylase (EC 3.2.1.2) (1
4-alpha-D-glucan maltohydrolase). [Alfalfa] {Medicago
sativa}, complete
Length = 1816
Score = 31.2 bits (69), Expect = 2.9
Identities = 28/87 (32%), Positives = 39/87 (44%), Gaps = 8/87 (9%)
Frame = -3
Query: 1289 VGNLTYIPRMSM-------SPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQTLDS-VGLY 1340
+ NLT PR ++ SP S W +R+ FP V Y + K+ ++DS LY
Sbjct: 890 IANLTLHPRKALLASSRI*SP*FSNWLEYQVRKNFPFSVRYVPFVLKN---SVDSGTSLY 720
Query: 1341 LPRSVFSHGQLYVAFSRVRTKAGLKIL 1367
+P S S AF+ K LK L
Sbjct: 719 VPASSGSSHSE*PAFAAAALKLSLKYL 639
>BQ135566
Length = 930
Score = 30.8 bits (68), Expect = 3.8
Identities = 24/96 (25%), Positives = 38/96 (39%), Gaps = 2/96 (2%)
Frame = +2
Query: 383 NPSWPEIQRFVGAKGLKPHDRPDIIARVFKIKFDELLNDLTKKHIL--GKVVAYMYTIEF 440
NPS P R + H DII + + LL + H + ++ Y Y +
Sbjct: 623 NPSPPYTSRLPLYINIYVHINHDIIYILSTTHYISLLYTIFHTHKITPNTIILYTYIPDP 802
Query: 441 QKRGLPHAHILIFLHPSSKYPTPDHINQIISAEIPH 476
P H I L+PS + P H++ + IP+
Sbjct: 803 ITYHTPSLHYHIILYPSYHFIIPIHVSPYTTLHIPY 910
>AJ500990 similar to PIR|T51278|T512 hypothetical protein T8M16_230 -
Arabidopsis thaliana, partial (5%)
Length = 376
Score = 30.4 bits (67), Expect = 5.0
Identities = 24/86 (27%), Positives = 37/86 (42%)
Frame = +3
Query: 10 SRESSNPEFGMCCGNGKIQIPFLPTPPPLLNDLMFNNYSTESKQFNQNIRLYNSMFAFSS 69
S + +N +G G +PF P PPPL D + +TE+ + RL
Sbjct: 99 SDDRANQTYGKLLGLVFSPVPFQPPPPPLETDDISEQKTTETTTTTASERLPR------K 260
Query: 70 PGFKVDKGVIPGRGPPTIRIQGQSCH 95
+ +GV+ R P + +QG S H
Sbjct: 261 EPVAIFQGVV--RLLPEVNLQGVSWH 332
>TC85667 similar to GP|15809788|gb|AAL06822.1 AT5g39740/MKM21_30 {Arabidopsis
thaliana}, partial (98%)
Length = 1189
Score = 30.4 bits (67), Expect = 5.0
Identities = 18/58 (31%), Positives = 30/58 (51%)
Frame = -2
Query: 1281 AKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQTLDSVG 1338
+ S N NL + +S PS++P FK + +FP++V T +++ LDS G
Sbjct: 615 SSFFSLSNPANLLSL*GISKPPSRAP--FKAPKTRFPVVVLTRPTSRRARNGLLDSAG 448
>TC81736 similar to SP|O49204|KAPS_CATRO Adenylylsulfate kinase chloroplast
precursor (EC 2.7.1.25) (APS kinase), partial (65%)
Length = 1033
Score = 30.4 bits (67), Expect = 5.0
Identities = 25/87 (28%), Positives = 39/87 (44%)
Frame = +2
Query: 929 QNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIASLLLPGGRTAHSKFKIP 988
Q G V +L G G+GK+ + L+ SL S+GK+ + +L G H +
Sbjct: 425 QKGCVIWLTGLSGSGKSTLACALSQSLHSRGKL----------TYILDGDNIRHGLNRDL 574
Query: 989 VPSFENSTCNIDGDSDLAKLLKVTNLI 1015
E+ + NI ++AKL LI
Sbjct: 575 SFRAEDRSENIRRIGEVAKLFADAGLI 655
>TC89058 similar to GP|19071218|gb|AAL84162.1 putative heavy metal
transporter {Arabidopsis thaliana}, partial (10%)
Length = 1618
Score = 30.0 bits (66), Expect = 6.5
Identities = 15/40 (37%), Positives = 26/40 (64%)
Frame = +1
Query: 131 NPNLNMQTVSKLQQMLDDTNCHAKSFRKARDRLRQGNVEN 170
NP+L + V+ L+ M + +CH S K+RD +++ N+EN
Sbjct: 574 NPSL-IGNVNPLKNMENHDHCHQGSCDKSRDGVQKHNIEN 690
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.336 0.147 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,938,361
Number of Sequences: 36976
Number of extensions: 619151
Number of successful extensions: 4417
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 2535
Number of HSP's successfully gapped in prelim test: 189
Number of HSP's that attempted gapping in prelim test: 1817
Number of HSP's gapped (non-prelim): 2847
length of query: 1391
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1283
effective length of database: 5,021,319
effective search space: 6442352277
effective search space used: 6442352277
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC148528.7


