

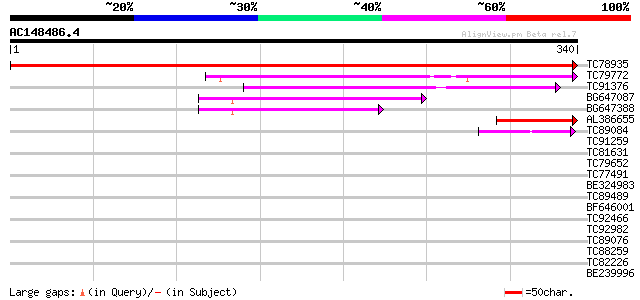
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.4 - phase: 0
(340 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78935 similar to GP|6760451|gb|AAF28357.1| S-ribonuclease bind... 689 0.0
TC79772 weakly similar to GP|20259023|gb|AAM14227.1 unknown prot... 127 8e-30
TC91376 weakly similar to GP|17065054|gb|AAL32681.1 putative pro... 96 3e-20
BG647087 weakly similar to GP|22136652|gb unknown protein {Arabi... 83 1e-16
BG647388 weakly similar to GP|22136652|gb| unknown protein {Arab... 65 5e-11
AL386655 weakly similar to GP|20259023|gb| unknown protein {Arab... 55 4e-08
TC89084 weakly similar to GP|9759369|dbj|BAB09828.1 gene_id:MYJ2... 44 1e-04
TC91259 weakly similar to GP|17065054|gb|AAL32681.1 putative pro... 40 0.002
TC81631 similar to GP|20279471|gb|AAM18751.1 unknown protein {Or... 37 0.011
TC79652 weakly similar to GP|19698951|gb|AAL91211.1 unknown prot... 36 0.024
TC77491 similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20 ... 36 0.024
BE324983 similar to PIR|T48442|T48 hypothetical protein T32M21.6... 36 0.024
TC89489 similar to PIR|T48442|T48442 hypothetical protein T32M21... 35 0.054
BF646001 homologue to GP|20303561|gb| putative kinase {Oryza sat... 33 0.12
TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 33 0.12
TC92982 similar to GP|23332794|gb|AAN27343.1 Sequence 13 from pa... 33 0.16
TC89076 similar to GP|18481708|gb|AAL73530.1 hypothetical protei... 32 0.35
TC88259 similar to GP|6729037|gb|AAF27033.1| unknown protein {Ar... 32 0.35
TC82226 similar to PIR|T48615|T48615 hypothetical protein F18O22... 32 0.46
BE239996 similar to GP|21689819|gb| unknown protein {Arabidopsis... 31 0.60
>TC78935 similar to GP|6760451|gb|AAF28357.1| S-ribonuclease binding protein
SBP1 {Petunia x hybrida}, partial (93%)
Length = 1304
Score = 689 bits (1779), Expect = 0.0
Identities = 340/340 (100%), Positives = 340/340 (100%)
Frame = +1
Query: 1 MAFLQDQFQRHYQQQQQPQPQTKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPF 60
MAFLQDQFQRHYQQQQQPQPQTKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPF
Sbjct: 58 MAFLQDQFQRHYQQQQQPQPQTKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPF 237
Query: 61 HVVGFAPGPVIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPRSVST 120
HVVGFAPGPVIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPRSVST
Sbjct: 238 HVVGFAPGPVIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPRSVST 417
Query: 121 GLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQAT 180
GLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQAT
Sbjct: 418 GLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQAT 597
Query: 181 QLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAA 240
QLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAA
Sbjct: 598 QLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAA 777
Query: 241 LKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVN 300
LKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVN
Sbjct: 778 LKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVN 957
Query: 301 EVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
EVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM
Sbjct: 958 EVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 1077
>TC79772 weakly similar to GP|20259023|gb|AAM14227.1 unknown protein
{Arabidopsis thaliana}, partial (33%)
Length = 1480
Score = 127 bits (318), Expect = 8e-30
Identities = 77/236 (32%), Positives = 124/236 (51%), Gaps = 13/236 (5%)
Frame = +2
Query: 118 VSTGLGLS--------LDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQL 169
VSTGLGLS L + S LSL+ + + +++QQ E+D+FL+ QGE+L
Sbjct: 569 VSTGLGLSFGDQQHQRLQLLQQQQCHSSHFLSLLSNGLASQIKQQKDEIDQFLQAQGEEL 748
Query: 170 RQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQ 229
++TI EK Q + E+ V ++LREKE +++ +RN ELE + L EA WQ
Sbjct: 749 QRTIEEKRQRNYRAIIKTAEETVARRLREKEIDLQKATRRNAELEARAAHLRTEAQLWQA 928
Query: 230 RARYNENMIAALKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNG-----RSLDFHLLSNE 284
+A+ E +L+ L A + G E G++E SC G + + E
Sbjct: 929 KAKEQEATAISLQTQLHHAMMSG--GAENRGENECG--LSCALGVEGHAEDAESGYIDPE 1096
Query: 285 NSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
+ + KC+ C ++V+LPC+HLC+C +C+++ CP+C + K +EVY+
Sbjct: 1097RAVVGSGPKCRGCGERVASVVVLPCRHLCVCTECDTRFGVCPVCFTVKNSTVEVYL 1264
>TC91376 weakly similar to GP|17065054|gb|AAL32681.1 putative protein
{Arabidopsis thaliana}, partial (41%)
Length = 997
Score = 95.5 bits (236), Expect = 3e-20
Identities = 53/190 (27%), Positives = 98/190 (50%)
Frame = +1
Query: 141 SLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKE 200
S + ++ LQ Q E+D F+ E++R I E+ + I++ V +KL++KE
Sbjct: 412 SFVDQNLLYHLQNQQSEIDLFIAQHTERVRMEIEEQRLKQSRMLQAAIQEAVTKKLKQKE 591
Query: 201 TEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGCG 260
E++ + K+N+ L+++ + L +E W++ A NE+ + L+ L+Q + +
Sbjct: 592 EEIQRMEKQNLMLQEKAKTLIMENQIWREMALTNESAVNTLRNELEQVLAHVENHR---- 759
Query: 261 DSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCES 320
++ D +SC + + ++ E S+ C C E ++LLPC+HLCLC C +
Sbjct: 760 -NDDDAASSCGSNHHVKEEVVVEEASSPVVGKLCSGCGERESVVLLLPCRHLCLCTMCGT 936
Query: 321 KLSFCPLCQS 330
+ CPLC S
Sbjct: 937 HIRNCPLCFS 966
>BG647087 weakly similar to GP|22136652|gb unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 718
Score = 83.2 bits (204), Expect = 1e-16
Identities = 51/152 (33%), Positives = 82/152 (53%), Gaps = 15/152 (9%)
Frame = +1
Query: 114 QPRSVSTGLGLSLDNTRLA---------------STGDSALLSLIGDDIDRELQQQDLEM 158
Q ++VSTGL LS D+ + S+ SL+ + +++QQ E+
Sbjct: 259 QQQNVSTGLRLSFDDQQQQRLQLQLQLQLHQQQQGCHSSSFSSLLPQGLVSQIKQQHDEL 438
Query: 159 DRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQME 218
D++L+ QGE LR+T+ +K Q + ++ E+ V ++LREKE E +RN ELE +
Sbjct: 439 DQYLQTQGENLRRTLADKRQKHYRELLNAAEEAVARRLREKEVEFAKATRRNAELEARTA 618
Query: 219 QLSVEAGAWQQRARYNENMIAALKFNLQQAYL 250
QL++EA WQ +A E A+L+ LQQ +
Sbjct: 619 QLTMEAQVWQAKAPAQEAAAASLQAQLQQTIM 714
>BG647388 weakly similar to GP|22136652|gb| unknown protein {Arabidopsis
thaliana}, partial (26%)
Length = 844
Score = 64.7 bits (156), Expect = 5e-11
Identities = 40/126 (31%), Positives = 68/126 (53%), Gaps = 15/126 (11%)
Frame = +3
Query: 114 QPRSVSTGLGLSLDNTRLA---------------STGDSALLSLIGDDIDRELQQQDLEM 158
Q ++VSTGL LS D+ + S+ SL+ + +++QQ E+
Sbjct: 420 QQQNVSTGLRLSFDDQQQQRLQLQLQLQLHQQQQGCHSSSFSSLLPQGLVSQIKQQHDEL 599
Query: 159 DRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQME 218
D++L+ QGE LR+T+ +K Q + ++ E+ V ++LREKE E + N ELE +
Sbjct: 600 DQYLQTQGENLRRTLADKRQKHYRELLNAAEEAVARRLREKEVEFAKATRTNAELEARTA 779
Query: 219 QLSVEA 224
QL++E+
Sbjct: 780 QLTMES 797
>AL386655 weakly similar to GP|20259023|gb| unknown protein {Arabidopsis
thaliana}, partial (20%)
Length = 498
Score = 55.1 bits (131), Expect = 4e-08
Identities = 19/48 (39%), Positives = 32/48 (66%)
Frame = +1
Query: 293 KCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
KC+ C T+V+LPC+HLC+C +C++ CP+C + K +EV++
Sbjct: 139 KCRGCEKRVATVVVLPCRHLCVCTECDAHFRVCPVCFTPKNSTVEVFL 282
>TC89084 weakly similar to GP|9759369|dbj|BAB09828.1
gene_id:MYJ24.10~ref|NP_055178.1~similar to unknown
protein {Arabidopsis thaliana}, partial (3%)
Length = 1200
Score = 43.5 bits (101), Expect = 1e-04
Identities = 18/58 (31%), Positives = 31/58 (53%)
Frame = +1
Query: 282 SNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
+ E K C+ C +EV + ++PC H+ LC+ C S +S CP C+ S + ++
Sbjct: 1021 TREADTAKAAWTCRVCLSSEVDITIVPCGHV-LCRKCSSAVSKCPFCRLSSTKAIRIF 1191
>TC91259 weakly similar to GP|17065054|gb|AAL32681.1 putative protein
{Arabidopsis thaliana}, partial (6%)
Length = 723
Score = 39.7 bits (91), Expect = 0.002
Identities = 50/192 (26%), Positives = 79/192 (41%)
Frame = +2
Query: 9 QRHYQQQQQPQPQTKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPFHVVGFAPG 68
Q HYQQQQQ Q N+ QM A PT + H +P F
Sbjct: 269 QHHYQQQQQQQMLNSELYNV-----QMDSASAVPLPT-----TMHESMLP------FYQS 400
Query: 69 PVIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPRSVSTGLGLSLDN 128
V + +D G L +N L+ +R R DF ++++ S L P
Sbjct: 401 NVCDPNRADSG--LTYNNPLQRKRSR----DF---STELVS---LPPH----------QK 514
Query: 129 TRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSII 188
R+ S+ S S GD + + Q Q ++DR L E++R + E+ +I
Sbjct: 515 NRVISSESS---SSFGDQVLYQFQNQQSDIDRILAHHNEKVRMELEEQKLRQSRMLXCMI 685
Query: 189 EDKVLQKLREKE 200
++ + +KL+EK+
Sbjct: 686 QETIAKKLKEKD 721
>TC81631 similar to GP|20279471|gb|AAM18751.1 unknown protein {Oryza sativa
(japonica cultivar-group)}, partial (40%)
Length = 787
Score = 37.0 bits (84), Expect = 0.011
Identities = 17/61 (27%), Positives = 28/61 (45%), Gaps = 4/61 (6%)
Frame = +1
Query: 273 GRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSF----CPLC 328
G S++ + NE +C C ++ PC+H+C+C C L F CP+C
Sbjct: 373 GNSVESDVDDNEQGK-----ECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPIC 537
Query: 329 Q 329
+
Sbjct: 538 R 540
>TC79652 weakly similar to GP|19698951|gb|AAL91211.1 unknown protein
{Arabidopsis thaliana}, partial (20%)
Length = 1590
Score = 35.8 bits (81), Expect = 0.024
Identities = 48/230 (20%), Positives = 86/230 (36%), Gaps = 41/230 (17%)
Frame = +2
Query: 140 LSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVL-QKLRE 198
+ IG D ++Q Q+ + + + + + +LE V+A Q Q I+E +++ Q++++
Sbjct: 185 IETIGQAYD-DMQTQNQHLLHQITERDDYNIKLVLEGVRARQKQDSFIMEMRLMEQEMQQ 361
Query: 199 KETEVENINKRNMELEDQME--------------QLSVEAGAWQQRA---RYNENMIAAL 241
+ N + ++EDQM Q SV+ Q+R R + +
Sbjct: 362 SNVSLNLYNTKAAKIEDQMRFCSDQIQKLVDNKLQSSVDLENTQRRLSDIRPSSQQVRNT 541
Query: 242 KFNLQQAYLQGRDS---------KEGCGDSEVDDTASCCNGR----------SLDFHLLS 282
+Q R + KE V+ S + L
Sbjct: 542 VVEVQSKITSSRVTHMELLVDLEKERFAKKRVEKDLEVARRNFSHLKAQDEDSSETDKLQ 721
Query: 283 NENSNMKDLMKCKACRVNEVTMVLLPCKHL----CLCKDCESKLSFCPLC 328
E +D++KC CR +V+ C HL C+ K S+ CP C
Sbjct: 722 QELGEYRDIVKCSICRDRTKEVVITKCYHLFCNSCIQKIAGSRQRKCPQC 871
>TC77491 similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20 {Arabidopsis
thaliana}, partial (81%)
Length = 1537
Score = 35.8 bits (81), Expect = 0.024
Identities = 15/43 (34%), Positives = 26/43 (59%)
Frame = +2
Query: 287 NMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
+ ++ M+C C N+ + C H+ C+DC S+LS CP+C+
Sbjct: 1106 DQRNQMECAICLTNKKDLAF-GCGHMT-CRDCGSRLSDCPICR 1228
>BE324983 similar to PIR|T48442|T48 hypothetical protein T32M21.60 -
Arabidopsis thaliana, partial (13%)
Length = 676
Score = 35.8 bits (81), Expect = 0.024
Identities = 15/41 (36%), Positives = 21/41 (50%), Gaps = 4/41 (9%)
Frame = +1
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKL----SFCPLCQS 330
C C N + +L C H+C C C S+L CPLC++
Sbjct: 298 CCVCCDNHIDSLLYRCGHMCTCSKCASELIRGGGKCPLCRA 420
>TC89489 similar to PIR|T48442|T48442 hypothetical protein T32M21.60 -
Arabidopsis thaliana, partial (5%)
Length = 1036
Score = 34.7 bits (78), Expect = 0.054
Identities = 29/125 (23%), Positives = 51/125 (40%), Gaps = 9/125 (7%)
Frame = +2
Query: 215 DQMEQLSVEAGAWQQRARYNENMIAA---LKFNLQQAYLQGRDSKEGCGDSEVDDTASCC 271
D + L ++ QQR + M+ A ++ LQ++ Q EV +
Sbjct: 245 DNISDLRIDMVRLQQRMNNMQRMLEACMDMQLELQRSIRQ-----------EVSAALNRS 391
Query: 272 NGRSLDFHLLSNENSNMKDLMK--CKACRVNEVTMVLLPCKHLCLCKDCESKL----SFC 325
G S +S + S + + K C C + +L C H+C C C ++L C
Sbjct: 392 TGSSGVHDHVSPDESKWECVRKGLCCICCEGSIDCLLYRCGHMCTCSKCANQLLDSRKKC 571
Query: 326 PLCQS 330
P+C++
Sbjct: 572 PMCRA 586
>BF646001 homologue to GP|20303561|gb| putative kinase {Oryza sativa
(japonica cultivar-group)}, partial (6%)
Length = 667
Score = 33.5 bits (75), Expect = 0.12
Identities = 24/87 (27%), Positives = 46/87 (52%)
Frame = +2
Query: 149 RELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINK 208
+EL+Q+++EM+ +K++ + R ++E + + IE+++ +K+ EKE EVE
Sbjct: 395 KELKQENVEME--MKVRELERRIGVIEMKEVEENSKRVRIEEEMKEKIYEKEMEVE---- 556
Query: 209 RNMELEDQMEQLSVEAGAWQQRARYNE 235
EL+ + VE W + R E
Sbjct: 557 ---ELKSMLMGKKVEEEKWLKDKRVLE 628
>TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (14%)
Length = 632
Score = 33.5 bits (75), Expect = 0.12
Identities = 17/48 (35%), Positives = 22/48 (45%)
Frame = +1
Query: 5 QDQFQRHYQQQQQPQPQTKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQ 52
++Q Q+ QQQQQPQPQ Q + QQ+ Q Q Q
Sbjct: 367 REQQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQ 510
Score = 28.5 bits (62), Expect = 3.9
Identities = 18/53 (33%), Positives = 27/53 (49%), Gaps = 3/53 (5%)
Frame = +1
Query: 4 LQDQFQRHYQQQQQPQPQTKSFRNLQTIEG---QMSQQMAFYNPTDLQDQSQH 53
L+ + Q+ QQQQQ QPQ + ++ Q + QM Q + + Q Q QH
Sbjct: 358 LKAREQQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQH 516
>TC92982 similar to GP|23332794|gb|AAN27343.1 Sequence 13 from patent US
6440698, partial (1%)
Length = 795
Score = 33.1 bits (74), Expect = 0.16
Identities = 26/77 (33%), Positives = 31/77 (39%)
Frame = -1
Query: 3 FLQDQFQRHYQQQQQPQPQTKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPFHV 62
+ Q Q Q+ QQQQQ QPQ QT + Q +Q Y P Q Q HP +
Sbjct: 618 YSQQQAQQAQQQQQQQQPQ-------QTQQTQQTQSYGSYPP---QAQPSHPEQHSWYGF 469
Query: 63 VGFAPGPVIPADGSDGG 79
AP S GG
Sbjct: 468 GSNAPSSFTSGGNSAGG 418
>TC89076 similar to GP|18481708|gb|AAL73530.1 hypothetical protein {Sorghum
bicolor}, partial (65%)
Length = 1326
Score = 32.0 bits (71), Expect = 0.35
Identities = 15/46 (32%), Positives = 22/46 (47%)
Frame = +2
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
C C N + C H+ C+DC S+L CP+C+ + VY
Sbjct: 1043 CPVCLTNAKDLAF-GCGHMT-CRDCGSRLRHCPICRHRITSRLRVY 1174
>TC88259 similar to GP|6729037|gb|AAF27033.1| unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1757
Score = 32.0 bits (71), Expect = 0.35
Identities = 25/97 (25%), Positives = 48/97 (48%), Gaps = 8/97 (8%)
Frame = +3
Query: 190 DKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNL---- 245
+ +++K E E ++E + +++E + + ++ + R R E + L+ L
Sbjct: 1362 EAMIEKNAELEKKLEKMEAAKLDVEMSLTECQMQLEKSENRIRAAELKVEELQTQLALAN 1541
Query: 246 ---QQAYLQGRDSKEGCGDSEVDDTASCCN-GRSLDF 278
Q+AY + +++K GDS V +T C N GR DF
Sbjct: 1542 KSNQEAYEELKETKTKKGDS*V-ETQICSN*GRRTDF 1649
>TC82226 similar to PIR|T48615|T48615 hypothetical protein F18O22.210 -
Arabidopsis thaliana, partial (63%)
Length = 1300
Score = 31.6 bits (70), Expect = 0.46
Identities = 20/66 (30%), Positives = 28/66 (42%), Gaps = 8/66 (12%)
Frame = +3
Query: 282 SNENS--------NMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKF 333
SNEN + D C C N M C H C +C L FCP+C+S+
Sbjct: 924 SNENEMSTEPSSGGLYDNKVCPICLTNAKDMAF-GCGHQTCC-ECGESLEFCPICRSTID 1097
Query: 334 IGMEVY 339
+++Y
Sbjct: 1098TRIKLY 1115
>BE239996 similar to GP|21689819|gb| unknown protein {Arabidopsis thaliana},
partial (16%)
Length = 420
Score = 31.2 bits (69), Expect = 0.60
Identities = 15/46 (32%), Positives = 21/46 (45%)
Frame = +2
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
C C N M C H CK+C + LS CP+C+ + +Y
Sbjct: 224 CPICLTNPKDMAF-GCGHTT-CKECGATLSLCPMCRQQITTRLRLY 355
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,769,740
Number of Sequences: 36976
Number of extensions: 141842
Number of successful extensions: 1202
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 1154
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1183
length of query: 340
length of database: 9,014,727
effective HSP length: 97
effective length of query: 243
effective length of database: 5,428,055
effective search space: 1319017365
effective search space used: 1319017365
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148486.4


