

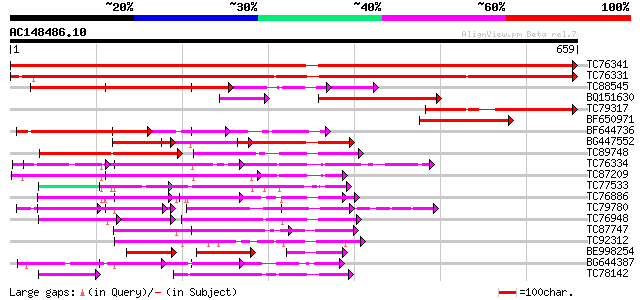
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.10 - phase: 0
(659 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76341 similar to GP|7528270|gb|AAF63202.1| poly(A)-binding pro... 1285 0.0
TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding pro... 976 0.0
TC88545 similar to GP|13560783|gb|AAK30205.1 poly(A)-binding pro... 394 e-110
BQ151630 similar to GP|7673359|gb| poly(A)-binding protein {Nico... 254 6e-68
TC79317 similar to GP|7673359|gb|AAF66825.1| poly(A)-binding pro... 226 2e-59
BF650971 similar to GP|7528270|gb|A poly(A)-binding protein {Cuc... 224 1e-58
BF644736 similar to GP|7528270|gb| poly(A)-binding protein {Cucu... 209 4e-54
BG447552 similar to GP|13560783|gb poly(A)-binding protein {Dauc... 182 4e-46
TC89748 homologue to PIR|B84565|B84565 probable spliceosome asso... 117 1e-26
TC76334 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein... 107 2e-23
TC87209 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein... 105 6e-23
TC77533 similar to GP|6996560|emb|CAB75429.1 oligouridylate bind... 89 4e-18
TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog... 87 2e-17
TC79780 similar to GP|18377731|gb|AAL67015.1 putative DNA bindin... 84 2e-16
TC76948 similar to SP|Q08935|ROC1_NICSY 29 kDa ribonucleoprotein... 78 9e-15
TC87747 weakly similar to PIR|C84768|C84768 hypothetical protein... 77 3e-14
TC92312 weakly similar to GP|4567275|gb|AAD23688.1| expressed pr... 75 8e-14
BE998254 weakly similar to GP|14029147|gb| polyadenylated mRNA-b... 74 1e-13
BG644387 homologue to GP|6996560|emb oligouridylate binding prot... 73 4e-13
TC78142 similar to GP|7578881|gb|AAF64167.1| plastid-specific ri... 72 5e-13
>TC76341 similar to GP|7528270|gb|AAF63202.1| poly(A)-binding protein
{Cucumis sativus}, partial (90%)
Length = 2501
Score = 1285 bits (3324), Expect = 0.0
Identities = 646/659 (98%), Positives = 647/659 (98%)
Frame = +3
Query: 1 MAQIQVQHQTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVS 60
MAQIQVQHQTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVS
Sbjct: 201 MAQIQVQHQTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVS 380
Query: 61 VRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTA 120
VRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTA
Sbjct: 381 VRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTA 560
Query: 121 NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKL 180
NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKL
Sbjct: 561 NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKL 740
Query: 181 NGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSA 240
NGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSA
Sbjct: 741 NGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSA 920
Query: 241 VLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRF 300
VLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDD+EWYVGKAQKKSEREQELKGRF
Sbjct: 921 VLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDKEWYVGKAQKKSEREQELKGRF 1100
Query: 301 EQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMR 360
EQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYK IMR
Sbjct: 1101EQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYK------------IMR 1244
Query: 361 DPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMR 420
DPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMR
Sbjct: 1245DPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMR 1424
Query: 421 PVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMP 480
PVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMP
Sbjct: 1425PVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMP 1604
Query: 481 SYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQDAPVQ 540
SYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQDAPVQ
Sbjct: 1605SYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQDAPVQ 1784
Query: 541 NIGGGMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQQRTMLGEALYPLVDQLEHDSA 600
NIGGGMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQQRTMLGEALYPLVDQLEHDSA
Sbjct: 1785NIGGGMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQQRTMLGEALYPLVDQLEHDSA 1964
Query: 601 AKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLASLSLNDS 659
AKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLASLSLNDS
Sbjct: 1965AKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLASLSLNDS 2141
>TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding protein
{Nicotiana tabacum}, partial (97%)
Length = 2581
Score = 976 bits (2522), Expect = 0.0
Identities = 493/667 (73%), Positives = 564/667 (83%), Gaps = 8/667 (1%)
Frame = +3
Query: 1 MAQIQVQHQTPAPVPAPSNGVVPNVA----NQFVTTSLYVGDLEVNVNDSQLYDLFNQVG 56
MAQ+QVQ Q + P+ G A FVTTSLYVGDL++NV DSQLYDLFNQ+G
Sbjct: 345 MAQVQVQPQNA--MTGPNGGAAAAAAAGGGGNFVTTSLYVGDLDMNVTDSQLYDLFNQLG 518
Query: 57 QVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRK 116
QVVSVRVCRDL TRRSLGYGYVN++NPQDAARALDVLNFTP+NN+ IR+MYSHRDPS RK
Sbjct: 519 QVVSVRVCRDLTTRRSLGYGYVNYSNPQDAARALDVLNFTPLNNRPIRIMYSHRDPSIRK 698
Query: 117 SGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNA 176
SG NIFIKNLDK IDHKALHDTFSSFG I+SCK+A DGSGQSKGYGFVQF+ E++AQ A
Sbjct: 699 SGQGNIFIKNLDKAIDHKALHDTFSSFGNILSCKVAVDGSGQSKGYGFVQFDTEEAAQKA 878
Query: 177 IDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGT 236
I+KLNGML+NDKQV+VG FLRKQ+R++ + KFNNV+VKNLSES T+D+LK FG +GT
Sbjct: 879 IEKLNGMLLNDKQVYVGPFLRKQERESTGDRAKFNNVFVKNLSESTTDDELKKTFGEFGT 1058
Query: 237 ITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQEL 296
ITSAV+MRD DG+SKCFGFVNFE+ +DAA+AVEALNGKK+DD+EWYVGKAQKKSERE EL
Sbjct: 1059ITSAVVMRDGDGKSKCFGFVNFESTDDAARAVEALNGKKIDDKEWYVGKAQKKSEREHEL 1238
Query: 297 KGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIR 356
K +FEQ++KE+ DK+QG NLY+KNLDDSI DEKLKE+FS +GTITS KV
Sbjct: 1239KIKFEQSMKEA-ADKYQGANLYVKNLDDSIADEKLKELFSSYGTITSCKV---------- 1385
Query: 357 VIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQF 416
MRDPNGVSRGSGFVAFSTPEEASRAL EMNGKM+ SKPLYV +AQRKEDRRARLQAQF
Sbjct: 1386--MRDPNGVSRGSGFVAFSTPEEASRALLEMNGKMVASKPLYVTLAQRKEDRRARLQAQF 1559
Query: 417 SQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGG 476
+QMRPV++ PSVAPRMP+YPPG PG+GQQ YGQGPPA++P Q GFGYQQQLVPGMRPGG
Sbjct: 1560AQMRPVSMPPSVAPRMPMYPPGGPGMGQQIFYGQGPPAIIPSQPGFGYQQQLVPGMRPGG 1739
Query: 477 GPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQD 536
P+P++FVPMVQQGQQGQRPGGRRGG QQ Q VP+M QQMLPR RVYRYPPGR + D
Sbjct: 1740APVPNFFVPMVQQGQQGQRPGGRRGGGVQQSQQPVPLMPQQMLPRGRVYRYPPGR-GMPD 1916
Query: 537 APVQNIGGGMMS--YDMGGLPLRD--VVPPMPIHALATALANAPPEQQRTMLGEALYPLV 592
P+ + GGM S YD+GG+P+RD + +PI ALA+ LANA PEQQRTMLGE LYPLV
Sbjct: 1917GPMPGVAGGMYSVPYDVGGMPIRDASLSQQIPIGALASHLANASPEQQRTMLGENLYPLV 2096
Query: 593 DQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLA 652
+QLE D+AAKVTGMLLEMDQ EVLHL+ESP+ALKAKVAEAM+VLRNV+QQQ ADQL+
Sbjct: 2097EQLEPDNAAKVTGMLLEMDQTEVLHLLESPEALKAKVAEAMDVLRNVAQQQAGGAADQLS 2276
Query: 653 SLSLNDS 659
SLSLNDS
Sbjct: 2277SLSLNDS 2297
>TC88545 similar to GP|13560783|gb|AAK30205.1 poly(A)-binding protein
{Daucus carota}, partial (35%)
Length = 866
Score = 394 bits (1011), Expect = e-110
Identities = 189/236 (80%), Positives = 211/236 (89%)
Frame = +2
Query: 25 VANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQ 84
V NQ TTSLYVGDL+ +V DSQLYDLFNQ+GQVVSVR+CRDLA+++SLGYGYVNF+NP
Sbjct: 158 VPNQLTTTSLYVGDLDHDVTDSQLYDLFNQIGQVVSVRICRDLASQQSLGYGYVNFSNPH 337
Query: 85 DAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFG 144
DAA+A+DVLNFTP+NNK IR+MYSHRDPS RKSG ANIFIKNLD+ IDHKAL+DTFS FG
Sbjct: 338 DAAKAMDVLNFTPLNNKPIRIMYSHRDPSVRKSGAANIFIKNLDRAIDHKALYDTFSIFG 517
Query: 145 QIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNV 204
I+SCKIA D SG SKGYGFVQFE E+SAQ+AIDKLNGML+NDK V+VGHF RKQDRDN
Sbjct: 518 NILSCKIAMDASGLSKGYGFVQFENEESAQSAIDKLNGMLLNDKPVYVGHFQRKQDRDNA 697
Query: 205 LSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFEN 260
LS KFNNVYVKNLSES T+DDLKN FG YGTITSAV+MRD DG+SKCFGFVNFEN
Sbjct: 698 LSNAKFNNVYVKNLSESVTDDDLKNTFGEYGTITSAVVMRDVDGKSKCFGFVNFEN 865
Score = 152 bits (385), Expect = 3e-37
Identities = 93/264 (35%), Positives = 139/264 (52%), Gaps = 1/264 (0%)
Frame = +2
Query: 112 PSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATD-GSGQSKGYGFVQFEAE 170
P + T ++++ +LD + L+D F+ GQ++S +I D S QS GYG+V F
Sbjct: 155 PVPNQLTTTSLYVGDLDHDVTDSQLYDLFNQIGQVVSVRICRDLASQQSLGYGYVNFSNP 334
Query: 171 DSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNE 230
A A+D LN +N+K + + + RD + K+ N+++KNL + L +
Sbjct: 335 HDAAKAMDVLNFTPLNNKPIRIMY----SHRDPSVRKSGAANIFIKNLDRAIDHKALYDT 502
Query: 231 FGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKS 290
F +G I S + DA G SK +GFV FEN E A A++ LNG ++D+ YVG Q+K
Sbjct: 503 FSIFGNILSCKIAMDASGLSKGYGFVQFENEESAQSAIDKLNGMLLNDKPVYVGHFQRKQ 682
Query: 291 EREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMI 350
+R+ L KF N+Y+KNL +S+TD+ LK F E+GTITS
Sbjct: 683 DRDNALSNA-----------KFN--NVYVKNLSESVTDDDLKNTFGEYGTITS------- 802
Query: 351 LFTDIRVIMRDPNGVSRGSGFVAF 374
V+MRD +G S+ GFV F
Sbjct: 803 -----AVVMRDVDGKSKCFGFVNF 859
Score = 98.2 bits (243), Expect = 9e-21
Identities = 68/219 (31%), Positives = 109/219 (49%), Gaps = 2/219 (0%)
Frame = +2
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEA 270
++YV +L T+ L + F G + S + RD A +S +G+VNF N DAAKA++
Sbjct: 182 SLYVGDLDHDVTDSQLYDLFNQIGQVVSVRICRDLASQQSLGYGYVNFSNPHDAAKAMDV 361
Query: 271 LNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEK 330
LN ++++ R + ++ V K N+++KNLD +I +
Sbjct: 362 LNFTPLNNKPI-----------------RIMYSHRDPSVRKSGAANIFIKNLDRAIDHKA 490
Query: 331 LKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGK 390
L + FS FG I S K+ D +G+S+G GFV F E A A+ ++NG
Sbjct: 491 LYDTFSIFGNILSCKIAM------------DASGLSKGYGFVQFENEESAQSAIDKLNGM 634
Query: 391 MIVSKPLYVAVAQRKEDR-RARLQAQFSQMRPVAITPSV 428
++ KP+YV QRK+DR A A+F+ + ++ SV
Sbjct: 635 LLNDKPVYVGHFQRKQDRDNALSNAKFNNVYVKNLSESV 751
>BQ151630 similar to GP|7673359|gb| poly(A)-binding protein {Nicotiana
tabacum}, partial (30%)
Length = 526
Score = 254 bits (650), Expect = 6e-68
Identities = 119/143 (83%), Positives = 131/143 (91%)
Frame = +1
Query: 360 RDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQM 419
R PNGVSRGSGFVAFSTPEEASRAL EMNGKM+ SKPLYV +AQRKEDRRARLQAQF+QM
Sbjct: 94 RGPNGVSRGSGFVAFSTPEEASRALLEMNGKMVASKPLYVTLAQRKEDRRARLQAQFAQM 273
Query: 420 RPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPM 479
RPV++ PSVAPRMP+YPPG PG+GQQ YGQGPPA++P Q GFGYQQQLVPGMRPGG P+
Sbjct: 274 RPVSMPPSVAPRMPMYPPGGPGMGQQIFYGQGPPAIIPSQPGFGYQQQLVPGMRPGGAPV 453
Query: 480 PSYFVPMVQQGQQGQRPGGRRGG 502
P++FVPMV QGQQGQRPGGRRGG
Sbjct: 454 PNFFVPMVXQGQQGQRPGGRRGG 522
Score = 45.1 bits (105), Expect = 9e-05
Identities = 23/59 (38%), Positives = 35/59 (58%)
Frame = +1
Query: 244 RDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQ 302
R +G S+ GFV F E+A++A+ +NGK V + YV AQ+K +R L+ +F Q
Sbjct: 94 RGPNGVSRGSGFVAFSTPEEASRALLEMNGKMVASKPLYVTLAQRKEDRRARLQAQFAQ 270
Score = 37.7 bits (86), Expect = 0.014
Identities = 17/46 (36%), Positives = 28/46 (59%)
Frame = +1
Query: 156 SGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDR 201
+G S+G GFV F + A A+ ++NG ++ K ++V RK+DR
Sbjct: 103 NGVSRGSGFVAFSTPEEASRALLEMNGKMVASKPLYVTLAQRKEDR 240
Score = 28.9 bits (63), Expect = 6.6
Identities = 15/44 (34%), Positives = 25/44 (56%)
Frame = +1
Query: 72 SLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSR 115
S G G+V F+ P++A+RAL +N + +K + V + R R
Sbjct: 112 SRGSGFVAFSTPEEASRALLEMNGKMVASKPLYVTLAQRKEDRR 243
>TC79317 similar to GP|7673359|gb|AAF66825.1| poly(A)-binding protein
{Nicotiana tabacum}, partial (32%)
Length = 848
Score = 226 bits (576), Expect = 2e-59
Identities = 122/177 (68%), Positives = 135/177 (75%), Gaps = 1/177 (0%)
Frame = +3
Query: 484 VPMVQQGQQGQRPGGRRGG-PGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQDAPVQNI 542
+PMVQQGQQGQRPGG RG P QQ P +PMMQQQM PR RVYRYP GR N Q P+ +
Sbjct: 3 MPMVQQGQQGQRPGGLRGASPMQQLPHSLPMMQQQMHPRGRVYRYPTGR-NPQGIPMPGV 179
Query: 543 GGGMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQQRTMLGEALYPLVDQLEHDSAAK 602
GG P+ ALATALANAPPEQQRTMLGE LYPLV+++EH AAK
Sbjct: 180 AGGT-----------------PMQALATALANAPPEQQRTMLGEVLYPLVEKIEHAGAAK 308
Query: 603 VTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLASLSLNDS 659
VTGMLLEMDQPEVLHLIESP+ALK KVAEA++VLRNV+QQQ NSP DQLASLSLND+
Sbjct: 309 VTGMLLEMDQPEVLHLIESPEALKTKVAEAVDVLRNVAQQQSNSPTDQLASLSLNDN 479
>BF650971 similar to GP|7528270|gb|A poly(A)-binding protein {Cucumis
sativus}, partial (8%)
Length = 657
Score = 224 bits (570), Expect = 1e-58
Identities = 106/109 (97%), Positives = 106/109 (97%)
Frame = +3
Query: 477 GPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQD 536
GPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQD
Sbjct: 330 GPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQD 509
Query: 537 APVQNIGGGMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQQRTMLG 585
APVQNIGG MMSYDMGGLPLRDVVPPMPIHALATALANAPPEQ TMLG
Sbjct: 510 APVQNIGGXMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQXXTMLG 656
>BF644736 similar to GP|7528270|gb| poly(A)-binding protein {Cucumis
sativus}, partial (20%)
Length = 674
Score = 209 bits (531), Expect = 4e-54
Identities = 102/158 (64%), Positives = 124/158 (77%)
Frame = +1
Query: 9 QTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLA 68
Q+P V APS+ + F SLYVGDLE +VN+ QL+DLF+QV QVVS+RVCRD
Sbjct: 208 QSPTVV-APSSAAL--TGGPFANASLYVGDLEGSVNEGQLFDLFSQVAQVVSIRVCRDQT 378
Query: 69 TRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLD 128
R SLGY YVNFTN QDAA A ++LNFTP+N K IR+M+S RDPS RKSG+ N+FIKNLD
Sbjct: 379 RRSSLGYAYVNFTNAQDAANAKELLNFTPLNEKPIRIMFSQRDPSVRKSGSGNVFIKNLD 558
Query: 129 KTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQ 166
+IDHKALHDTF++FG ++SCK+A DG+GQSK YGFVQ
Sbjct: 559 TSIDHKALHDTFAAFGPVLSCKVALDGNGQSKXYGFVQ 672
Score = 74.7 bits (182), Expect = 1e-13
Identities = 42/138 (30%), Positives = 74/138 (53%), Gaps = 1/138 (0%)
Frame = +1
Query: 120 ANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSK-GYGFVQFEAEDSAQNAID 178
A++++ +L+ +++ L D FS Q++S ++ D + +S GY +V F A NA +
Sbjct: 268 ASLYVGDLEGSVNEGQLFDLFSQVAQVVSIRVCRDQTRRSSLGYAYVNFTNAQDAANAKE 447
Query: 179 KLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTIT 238
LN +N+K + + RD + K+ NV++KNL S L + F A+G +
Sbjct: 448 LLNFTPLNEKPIRIMF----SQRDPSVRKSGSGNVFIKNLDTSIDHKALHDTFAAFGPVL 615
Query: 239 SAVLMRDADGRSKCFGFV 256
S + D +G+SK +GFV
Sbjct: 616 SCKVALDGNGQSKXYGFV 669
Score = 60.8 bits (146), Expect = 2e-09
Identities = 50/162 (30%), Positives = 74/162 (44%), Gaps = 1/162 (0%)
Frame = +1
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKC-FGFVNFENAEDAAKAVEA 270
++YV +L S E L + F + S + RD RS + +VNF NA+DAA A E
Sbjct: 271 SLYVGDLEGSVNEGQLFDLFSQVAQVVSIRVCRDQTRRSSLGYAYVNFTNAQDAANAKEL 450
Query: 271 LNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEK 330
LN ++ E+ ++ F Q ++ V K N+++KNLD SI +
Sbjct: 451 LNFTPLN---------------EKPIRIMFSQ--RDPSVRKSGSGNVFIKNLDTSIDHKA 579
Query: 331 LKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFV 372
L + F+ FG + S KV D NG S+ GFV
Sbjct: 580 LHDTFAAFGPVLSCKVAL------------DGNGQSKXYGFV 669
>BG447552 similar to GP|13560783|gb poly(A)-binding protein {Daucus carota},
partial (18%)
Length = 372
Score = 182 bits (462), Expect = 4e-46
Identities = 94/136 (69%), Positives = 108/136 (79%)
Frame = +1
Query: 265 AKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDD 324
A+ VEALNGKK DD+EWYVGKA KK ERE ELK R EQ++KE+V DKF G NLYLKNLDD
Sbjct: 1 ARGVEALNGKKFDDKEWYVGKALKKYERELELKERHEQSMKETV-DKFYGANLYLKNLDD 177
Query: 325 SITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRAL 384
S+TDEKL E+FSEFGT+TS K I+RDP G+SRGSGFVAFSTPEEA++AL
Sbjct: 178 SVTDEKLSELFSEFGTVTSCK------------ILRDPQGISRGSGFVAFSTPEEATQAL 321
Query: 385 GEMNGKMIVSKPLYVA 400
EMNGKM+ KP+YVA
Sbjct: 322 AEMNGKMVAGKPVYVA 369
Score = 75.1 bits (183), Expect = 8e-14
Identities = 43/119 (36%), Positives = 67/119 (56%), Gaps = 12/119 (10%)
Frame = +1
Query: 177 IDKLNGMLINDKQVFVGHFLRKQDRDNVLSKT----------KFN--NVYVKNLSESFTE 224
++ LNG +DK+ +VG L+K +R+ L + KF N+Y+KNL +S T+
Sbjct: 10 VEALNGKKFDDKEWYVGKALKKYERELELKERHEQSMKETVDKFYGANLYLKNLDDSVTD 189
Query: 225 DDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYV 283
+ L F +GT+TS ++RD G S+ GFV F E+A +A+ +NGK V + YV
Sbjct: 190 EKLSELFSEFGTVTSCKILRDPQGISRGSGFVAFSTPEEATQALAEMNGKMVAGKPVYV 366
Score = 67.4 bits (163), Expect = 2e-11
Identities = 30/73 (41%), Positives = 46/73 (62%)
Frame = +1
Query: 120 ANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDK 179
AN+++KNLD ++ + L + FS FG + SCKI D G S+G GFV F + A A+ +
Sbjct: 148 ANLYLKNLDDSVTDEKLSELFSEFGTVTSCKILRDPQGISRGSGFVAFSTPEEATQALAE 327
Query: 180 LNGMLINDKQVFV 192
+NG ++ K V+V
Sbjct: 328 MNGKMVAGKPVYV 366
>TC89748 homologue to PIR|B84565|B84565 probable spliceosome associated
protein [imported] - Arabidopsis thaliana, partial (56%)
Length = 707
Score = 117 bits (294), Expect = 1e-26
Identities = 64/169 (37%), Positives = 107/169 (62%), Gaps = 2/169 (1%)
Frame = +2
Query: 35 YVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLN 94
YVG+L+ +++ L++LF Q G VV+V V +D T + GYG+V F + +DA A+ VLN
Sbjct: 173 YVGNLDPQISEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEFRSEEDADYAIKVLN 352
Query: 95 FTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIM-SCKIAT 153
+ K IRV + +D S G AN+FI NLD +D K L+DTFS+FG I+ + KI
Sbjct: 353 MIKLYGKPIRVNKASQDKKSLDVG-ANLFIGNLDPDVDEKLLYDTFSAFGVIVTNPKIMR 529
Query: 154 D-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDR 201
D + S+G+GF+ +++ +++ +AI+ +NG + ++Q+ V + +K +
Sbjct: 530 DPDTRNSRGFGFISYDSFEASDSAIEAMNGQYLCNRQITVSYAYKKDTK 676
Score = 69.7 bits (169), Expect = 3e-12
Identities = 56/200 (28%), Positives = 96/200 (48%), Gaps = 2/200 (1%)
Frame = +2
Query: 214 YVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALN 272
YV NL +E+ L F G + + + +D + + +GFV F + EDA A++ LN
Sbjct: 173 YVGNLDPQISEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEFRSEEDADYAIKVLN 352
Query: 273 GKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLK 332
K+ + V KA S+ ++ L G NL++ NLD + ++ L
Sbjct: 353 MIKLYGKPIRVNKA---SQDKKSLD---------------VGANLFIGNLDPDVDEKLLY 478
Query: 333 EMFSEFGTITSYKVCTMILFTDIRVIMRDPNGV-SRGSGFVAFSTPEEASRALGEMNGKM 391
+ FS FG I + IMRDP+ SRG GF+++ + E + A+ MNG+
Sbjct: 479 DTFSAFGVIVTNPK-----------IMRDPDTRNSRGFGFISYDSFEASDSAIEAMNGQY 625
Query: 392 IVSKPLYVAVAQRKEDRRAR 411
+ ++ + V+ A +K+ + R
Sbjct: 626 LCNRQITVSYAYKKDTKGER 685
Score = 35.4 bits (80), Expect = 0.070
Identities = 25/95 (26%), Positives = 43/95 (44%)
Frame = +2
Query: 310 DKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGS 369
++ Q Y+ NLD I++E L E+F + G + ++ V +G
Sbjct: 149 ERNQDATAYVGNLDPQISEELLWELFVQAGPV-----------VNVYVPKDRVTNQHQGY 295
Query: 370 GFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQR 404
GFV F + E+A A+ +N + KP+ V A +
Sbjct: 296 GFVEFRSEEDADYAIKVLNMIKLYGKPIRVNKASQ 400
>TC76334 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein 45
{Nicotiana plumbaginifolia}, partial (68%)
Length = 1579
Score = 107 bits (266), Expect = 2e-23
Identities = 82/293 (27%), Positives = 142/293 (47%), Gaps = 23/293 (7%)
Frame = +1
Query: 4 IQVQHQTPAPVPAPSNGVVPNV-ANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVR 62
+ +Q Q P + A + P A+ +L++GDL+ ++++ LY F G++ SV+
Sbjct: 142 VMMQPQQPPAMWAAQSAQPPQQPASADEVRTLWIGDLQYWMDENYLYTCFGNTGELTSVK 321
Query: 63 VCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNN--KSIRV---MYSHRDPSSRKS 117
V R+ T +S GYG++ F A R L N T M N ++ R+ +S + SSR+
Sbjct: 322 VIRNKQTSQSEGYGFIEFNTRATAERVLQTYNGTIMPNGGQNYRLNWATFSAGERSSRQD 501
Query: 118 GTAN--IFIKNLDKTIDHKALHDTF-SSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSA 173
+ IF+ +L + L +TF + + + K+ D +G+SKGYGFV+F E
Sbjct: 502 DGPDHTIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRSKGYGFVRFADEGEQ 681
Query: 174 QNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFN-------------NVYVKNLSE 220
A+ ++ G+L + + + +G K K +N ++V NL
Sbjct: 682 MRAMTEMQGVLCSTRPMRIGPATNKNPAATTQPKASYNPSGGQSENDPNNTTIFVGNLDP 861
Query: 221 SFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNG 273
+ T+D L+ F YG + V ++ G+ +C GFV F + A +A+ LNG
Sbjct: 862 NVTDDHLRQVFTQYGEL---VHVKIPSGK-RC-GFVQFSDRSCAEEAIRVLNG 1005
Score = 84.3 bits (207), Expect = 1e-16
Identities = 87/379 (22%), Positives = 164/379 (42%), Gaps = 7/379 (1%)
Frame = +1
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
++I +L +D L+ F + G++ S K+ + + QS+GYGF++F +A+ +
Sbjct: 235 LWIGDLQYWMDENYLYTCFGNTGELTSVKVIRNKQTSQSEGYGFIEFNTRATAERVLQTY 414
Query: 181 NGMLI-NDKQVFVGHF--LRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGA-YGT 236
NG ++ N Q + ++ +R + + ++V +L+ T+ L+ F A Y +
Sbjct: 415 NGTIMPNGGQNYRLNWATFSAGERSSRQDDGPDHTIFVGDLAADVTDYLLQETFRARYNS 594
Query: 237 ITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQ- 294
+ A ++ D GRSK +GFV F + + +A+ + G +G A K+
Sbjct: 595 VKGAKVVIDRLTGRSKGYGFVRFADEGEQMRAMTEMQGVLCSTRPMRIGPATNKNPAATT 774
Query: 295 ELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTD 354
+ K + + +S D +++ NLD ++TD+ L+++F+++G + K+
Sbjct: 775 QPKASYNPSGGQSENDP-NNTTIFVGNLDPNVTDDHLRQVFTQYGELVHVKI-------- 927
Query: 355 IRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQA 414
P+G + GFV FS A A+ +NG ++ + + ++ + +++
Sbjct: 928 -------PSG--KRCGFVQFSDRSCAEEAIRVLNGTLLGGQNVRLSWGRTPSNKQ----- 1065
Query: 415 QFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRP 474
+Q P G P G + Y QG A Q V G P
Sbjct: 1066--TQQDPT-------------QGGYPAAGGYYGYAQGGYENYGYAAPPAGQDPNVYGSYP 1200
Query: 475 GGGPMPSYFVPMVQQGQQG 493
G P Y P QQ Q G
Sbjct: 1201G---YPGYQHPQQQQQQMG 1248
Score = 48.5 bits (114), Expect = 8e-06
Identities = 31/101 (30%), Positives = 55/101 (53%)
Frame = +1
Query: 17 PSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYG 76
PS G N N T+++VG+L+ NV D L +F Q G++V V++ G
Sbjct: 796 PSGGQSENDPNN---TTIFVGNLDPNVTDDHLRQVFTQYGELVHVKIPSGKRC------G 948
Query: 77 YVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKS 117
+V F++ A A+ VLN T + +++R+ + R PS++++
Sbjct: 949 FVQFSDRSCAEEAIRVLNGTLLGGQNVRLSWG-RTPSNKQT 1068
>TC87209 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein 45
{Nicotiana plumbaginifolia}, partial (69%)
Length = 1504
Score = 105 bits (262), Expect = 6e-23
Identities = 86/323 (26%), Positives = 152/323 (46%), Gaps = 32/323 (9%)
Frame = +3
Query: 3 QIQVQHQTPAP------VPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVG 56
Q Q Q QT P P VP ++ +L++GDL+ ++++ LY+ F+ G
Sbjct: 153 QPQPQPQTQPPQMWAPNAQPPQQSAVPPPSSADEVKTLWIGDLQYWMDENYLYNCFSHTG 332
Query: 57 QVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNN--KSIRV---MYSHRD 111
+V SV+V R+ T +S GYG++ F + A R L N T M N ++ R+ +S +
Sbjct: 333 EVGSVKVIRNKQTNQSEGYGFLEFISRAGAERVLQTFNGTIMPNGGQNFRLNWATFSSGE 512
Query: 112 PSSRKSGTANIFIKNLDKTIDHKALHDTF-SSFGQIMSCKIATD-GSGQSKGYGFVQFEA 169
S IF+ +L + L + F + + + K+ D +G++KGYGFV+F
Sbjct: 513 KRHDDSPDYTIFVGDLAADVSDHHLTEVFRTRYNSVKGAKVVIDRTTGRTKGYGFVRFAD 692
Query: 170 EDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNN---------------VY 214
E A+ ++ G+L + + + +G K + SK + N ++
Sbjct: 693 ESEQMRAMTEMQGVLCSTRPMRIGPASNK-NLGTQTSKASYQNPQGGAQNENDPNNTTIF 869
Query: 215 VKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGK 274
V NL + T++ LK F YG + V ++ G+ +C GFV F + A +A+ LNG
Sbjct: 870 VGNLDPNVTDEHLKQVFTQYGEL---VHVKIPSGK-RC-GFVQFADRSSAEEALRVLNGT 1034
Query: 275 KVDDE----EWYVGKAQKKSERE 293
+ + W A K+++++
Sbjct: 1035LLGGQNVRLSWGRSPANKQTQQD 1103
Score = 87.4 bits (215), Expect = 2e-17
Identities = 69/287 (24%), Positives = 133/287 (46%), Gaps = 6/287 (2%)
Frame = +3
Query: 112 PSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAE 170
P S ++I +L +D L++ FS G++ S K+ + + QS+GYGF++F +
Sbjct: 234 PPSSADEVKTLWIGDLQYWMDENYLYNCFSHTGEVGSVKVIRNKQTNQSEGYGFLEFISR 413
Query: 171 DSAQNAIDKLNGMLI-NDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKN 229
A+ + NG ++ N Q F ++ + + ++V +L+ ++ L
Sbjct: 414 AGAERVLQTFNGTIMPNGGQNFRLNWATFSSGEKRHDDSPDYTIFVGDLAADVSDHHLTE 593
Query: 230 EFGA-YGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQ 287
F Y ++ A ++ D GR+K +GFV F + + +A+ + G +G A
Sbjct: 594 VFRTRYNSVKGAKVVIDRTTGRTKGYGFVRFADESEQMRAMTEMQGVLCSTRPMRIGPAS 773
Query: 288 KKSEREQELKGRFE--QTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYK 345
K+ Q K ++ Q ++ D +++ NLD ++TDE LK++F+++G + K
Sbjct: 774 NKNLGTQTSKASYQNPQGGAQNENDP-NNTTIFVGNLDPNVTDEHLKQVFTQYGELVHVK 950
Query: 346 VCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMI 392
+ P+G + GFV F+ A AL +NG ++
Sbjct: 951 I---------------PSG--KRCGFVQFADRSSAEEALRVLNGTLL 1040
>TC77533 similar to GP|6996560|emb|CAB75429.1 oligouridylate binding protein
{Nicotiana plumbaginifolia}, partial (79%)
Length = 1621
Score = 89.4 bits (220), Expect = 4e-18
Identities = 82/310 (26%), Positives = 130/310 (41%), Gaps = 17/310 (5%)
Frame = +1
Query: 105 VMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGF 164
++ + P S ++++ N+ + L + FSS G + CK+ + YGF
Sbjct: 175 ILSGNLPPGFDSSTCRSVYVGNIHPQVSEPLLQELFSSAGALEGCKLIRK---EKSSYGF 345
Query: 165 VQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTE 224
V + SA AI LNG I + + V + R++ +++V +LS T+
Sbjct: 346 VDYFDRSSAAIAIVTLNGRNIFGQSIKVNWAYTRGQREDTSGHF---HIFVGDLSPEVTD 516
Query: 225 DDLKNEFGAYGTITSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDEE--- 280
L F AY + + A +M D GRS+ FGFV+F N ++A A+ L GK + +
Sbjct: 517 ATLYACFSAYSSCSDARVMWDQKTGRSRGFGFVSFRNQQEAQSAINDLTGKWLGSRQIRC 696
Query: 281 -WYVGKAQKKSEREQ-------ELKGRFEQTVKESVVDKF-----QGLNLYLKNLDDSIT 327
W A E + EL + +E D Q +Y+ NL +T
Sbjct: 697 NWATKGANMNGENQSSESKSVVELTSGTSEEAQEMTSDDSPEKNPQYTTVYVGNLAPEVT 876
Query: 328 DEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEM 387
L F G T D+RV RD +G GFV +ST EA+ A+
Sbjct: 877 SVDLHHHFHALGVGT---------IEDVRV-QRD-----KGFGFVRYSTHGEAALAIQMG 1011
Query: 388 NGKMIVSKPL 397
N + + KP+
Sbjct: 1012NTRFLFGKPI 1041
Score = 55.1 bits (131), Expect = 9e-08
Identities = 50/192 (26%), Positives = 76/192 (39%), Gaps = 35/192 (18%)
Frame = +1
Query: 34 LYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVL 93
++VGDL V D+ LY F+ RV D T RS G+G+V+F N Q+A A++ L
Sbjct: 481 IFVGDLSPEVTDATLYACFSAYSSCSDARVMWDQKTGRSRGFGFVSFRNQQEAQSAINDL 660
Query: 94 NFTPMNNKSIRV--------MYSHRDPSSRK------SGTA------------------- 120
+ ++ IR M S K SGT+
Sbjct: 661 TGKWLGSRQIRCNWATKGANMNGENQSSESKSVVELTSGTSEEAQEMTSDDSPEKNPQYT 840
Query: 121 NIFIKNLDKTIDHKALHDTFSSF--GQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAID 178
+++ NL + LH F + G I ++ D KG+GFV++ A AI
Sbjct: 841 TVYVGNLAPEVTSVDLHHHFHALGVGTIEDVRVQRD-----KGFGFVRYSTHGEAALAIQ 1005
Query: 179 KLNGMLINDKQV 190
N + K +
Sbjct: 1006MGNTRFLFGKPI 1041
>TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment), partial (67%)
Length = 1896
Score = 87.4 bits (215), Expect = 2e-17
Identities = 74/280 (26%), Positives = 131/280 (46%), Gaps = 39/280 (13%)
Frame = +2
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
++++GDL ++++ L++ F G+V S +V R+ T +S GYG+V F A + L
Sbjct: 551 TIWLGDLHHWMDETFLHNCFAHTGEVASAKVIRNKQTGQSEGYGFVEFYTRAMAEKVLQN 730
Query: 93 LNFTPMNN--KSIRVMYS--------HRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSS 142
N T M N ++ R+ ++ SS + ++F+ +L + L +TF+S
Sbjct: 731 FNGTMMPNTDQAFRLNWATFSAAGGGGERRSSEATSDLSVFVGDLAIDVTDAMLQETFAS 910
Query: 143 -FGQIMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQD 200
F I K+ D +G+SKGYGFV+F E A+ ++NG+ + + + VG K+
Sbjct: 911 KFSSIKGAKVVIDSNTGRSKGYGFVRFGDESERTRAMTEMNGVYCSSRPMRVGVATPKKT 1090
Query: 201 RDN---------VL----------------SKTKFNN--VYVKNLSESFTEDDLKNEFGA 233
N VL S+ NN ++V L +++DL+ F
Sbjct: 1091YGNPQQYSSQAVVLAGGGGHSNGAMAQGSQSEGDSNNTTIFVGGLDSDISDEDLRQPFLQ 1270
Query: 234 YGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNG 273
+G + S + K GFV + ++A +A++ LNG
Sbjct: 1271FGDVISVKI-----PVGKGCGFVQLADRKNAEEAIQGLNG 1375
Score = 84.0 bits (206), Expect = 2e-16
Identities = 79/305 (25%), Positives = 134/305 (43%), Gaps = 34/305 (11%)
Frame = +2
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
I++ +L +D LH+ F+ G++ S K+ + +GQS+GYGFV+F A+ +
Sbjct: 554 IWLGDLHHWMDETFLHNCFAHTGEVASAKVIRNKQTGQSEGYGFVEFYTRAMAEKVLQNF 733
Query: 181 NG-MLINDKQVFV-----------GHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLK 228
NG M+ N Q F G R + + LS V+V +L+ T+ L+
Sbjct: 734 NGTMMPNTDQAFRLNWATFSAAGGGGERRSSEATSDLS------VFVGDLAIDVTDAMLQ 895
Query: 229 NEFGA-YGTITSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKA 286
F + + +I A ++ D++ GRSK +GFV F + + +A+ +NG VG A
Sbjct: 896 ETFASKFSSIKGAKVVIDSNTGRSKGYGFVRFGDESERTRAMTEMNGVYCSSRPMRVGVA 1075
Query: 287 QKKSEREQELKGRFEQTVKESVVDKFQG-------------------LNLYLKNLDDSIT 327
K + G +Q ++VV G +++ LD I+
Sbjct: 1076TPK-----KTYGNPQQYSSQAVVLAGGGGHSNGAMAQGSQSEGDSNNTTIFVGGLDSDIS 1240
Query: 328 DEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEM 387
DE L++ F +FG + S K+ V +G GFV + + A A+ +
Sbjct: 1241DEDLRQPFLQFGDVISVKI-----------------PVGKGCGFVQLADRKNAEEAIQGL 1369
Query: 388 NGKMI 392
NG +I
Sbjct: 1370NGTVI 1384
Score = 71.2 bits (173), Expect = 1e-12
Identities = 54/190 (28%), Positives = 82/190 (42%), Gaps = 32/190 (16%)
Frame = +2
Query: 33 SLYVGDLEVNVNDSQLYDLF-NQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL ++V D+ L + F ++ + +V D T RS GYG+V F + + RA+
Sbjct: 845 SVFVGDLAIDVTDAMLQETFASKFSSIKGAKVVIDSNTGRSKGYGFVRFGDESERTRAMT 1024
Query: 92 VLNFTPMNNKSIRV-------------MYSHR------------------DPSSRKSGTA 120
+N +++ +RV YS + S S
Sbjct: 1025 EMNGVYCSSRPMRVGVATPKKTYGNPQQYSSQAVVLAGGGGHSNGAMAQGSQSEGDSNNT 1204
Query: 121 NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKL 180
IF+ LD I + L F FG ++S KI KG GFVQ +A+ AI L
Sbjct: 1205 TIFVGGLDSDISDEDLRQPFLQFGDVISVKIPV-----GKGCGFVQLADRKNAEEAIQGL 1369
Query: 181 NGMLINDKQV 190
NG +I + V
Sbjct: 1370 NGTVIGKQTV 1399
Score = 66.2 bits (160), Expect = 4e-11
Identities = 59/213 (27%), Positives = 100/213 (46%), Gaps = 4/213 (1%)
Frame = +2
Query: 198 KQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDAD-GRSKCFGFV 256
+Q + L+ + +++ +L E L N F G + SA ++R+ G+S+ +GFV
Sbjct: 509 QQHQQKQLAPEEIRTIWLGDLHHWMDETFLHNCFAHTGEVASAKVIRNKQTGQSEGYGFV 688
Query: 257 NFENAEDAAKAVEALNGKKVD--DEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQG 314
F A K ++ NG + D+ + + A + G E+ E+ D
Sbjct: 689 EFYTRAMAEKVLQNFNGTMMPNTDQAFRLNWATFSAAG-----GGGERRSSEATSD---- 841
Query: 315 LNLYLKNLDDSITDEKLKEMF-SEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVA 373
L++++ +L +TD L+E F S+F +I KV ++ G S+G GFV
Sbjct: 842 LSVFVGDLAIDVTDAMLQETFASKFSSIKGAKV-----------VIDSNTGRSKGYGFVR 988
Query: 374 FSTPEEASRALGEMNGKMIVSKPLYVAVAQRKE 406
F E +RA+ EMNG S+P+ V VA K+
Sbjct: 989 FGDESERTRAMTEMNGVYCSSRPMRVGVATPKK 1087
>TC79780 similar to GP|18377731|gb|AAL67015.1 putative DNA binding protein
ACBF {Arabidopsis thaliana}, partial (67%)
Length = 1649
Score = 84.0 bits (206), Expect = 2e-16
Identities = 47/158 (29%), Positives = 87/158 (54%), Gaps = 7/158 (4%)
Frame = +3
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+L++GDL+ V+++ L F+ G+V+S+++ R+ T + GYG++ F + A R L
Sbjct: 102 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 281
Query: 93 LNFT--PMNNKSIRVMYSHRDPSSRKSGTA---NIFIKNLDKTIDHKALHDTF-SSFGQI 146
N T P ++ R+ ++ R+ +IF+ +L + L +TF + +G +
Sbjct: 282 YNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSV 461
Query: 147 MSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGM 183
K+ TD +G+SKGYGFV+F E A+ ++NG+
Sbjct: 462 RGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGV 575
Score = 53.5 bits (127), Expect = 2e-07
Identities = 48/199 (24%), Positives = 91/199 (45%), Gaps = 4/199 (2%)
Frame = +3
Query: 206 SKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGRSKCFGFVNFENAEDA 264
S + +++ +L E+ L + F G + S ++R+ G+ + +GF+ F + A
Sbjct: 84 SLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAA 263
Query: 265 AKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQT-VKESVVDKFQGLNLYLKNLD 323
+ ++ NG ++ E Q + + + E D ++++ +L
Sbjct: 264 ERVLQTYNGTQMPGTE-------------QTFRLNWASFGIGERRPDAGPDHSIFVGDLA 404
Query: 324 DSITDEKLKEMF-SEFGTITSYKVCTMILFTDIRVIMRDPN-GVSRGSGFVAFSTPEEAS 381
+TD L+E F + +G++ KV T DPN G S+G GFV FS E +
Sbjct: 405 PDVTDYLLQETFRTHYGSVRGAKVVT------------DPNTGRSKGYGFVKFSDESERN 548
Query: 382 RALGEMNGKMIVSKPLYVA 400
RA+ EMNG ++P+ ++
Sbjct: 549 RAMSEMNGVYCSTRPMRIS 605
Score = 47.4 bits (111), Expect = 2e-05
Identities = 26/81 (32%), Positives = 39/81 (48%)
Frame = +2
Query: 112 PSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAED 171
P I++ NLD + + L F FG+I+S K+ K GFVQF A
Sbjct: 734 PPEYDVNNTTIYVGNLDLNVSEEELKQNFLQFGEIVSVKV-----HPGKACGFVQFGARA 898
Query: 172 SAQNAIDKLNGMLINDKQVFV 192
SA+ AI K+ G ++ + + V
Sbjct: 899 SAEEAIQKMQGKILGQQVIRV 961
Score = 45.4 bits (106), Expect = 7e-05
Identities = 44/182 (24%), Positives = 74/182 (40%)
Frame = +2
Query: 317 LYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFST 376
+Y+ NLD ++++E+LK+ F +FG I S KV + GFV F
Sbjct: 764 IYVGNLDLNVSEEELKQNFLQFGEIVSVKV-----------------HPGKACGFVQFGA 892
Query: 377 PEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYP 436
A A+ +M GK++ + + V+ R + R + + Q + Y
Sbjct: 893 RASAEEAIQKMQGKILGQQVIRVSWG-RPQTARQDVPGGWGQQ------VDQSQWSAYYG 1051
Query: 437 PGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMPSYFVPMVQQGQQGQRP 496
G PG + + YG M AG+ Q P G + + VP ++Q ++ P
Sbjct: 1052YGQPGY-EAYAYGAAQDPSMYAYAGYASYAQ-YPQQVEGAQDVSAMSVPTLEQREELYDP 1225
Query: 497 GG 498
G
Sbjct: 1226SG 1231
Score = 43.9 bits (102), Expect = 2e-04
Identities = 31/100 (31%), Positives = 49/100 (49%)
Frame = +2
Query: 8 HQTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDL 67
+ PA AP N V P T++YVG+L++NV++ +L F Q G++VSV+V
Sbjct: 692 YPVPAYTTAPVNTVPPEY--DVNNTTIYVGNLDLNVSEEELKQNFLQFGEIVSVKVHPGK 865
Query: 68 ATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMY 107
A G+V F A A+ + + + IRV +
Sbjct: 866 AC------GFVQFGARASAEEAIQKMQGKILGQQVIRVSW 967
>TC76948 similar to SP|Q08935|ROC1_NICSY 29 kDa ribonucleoprotein A
chloroplast precursor (CP29A). [Wood tobacco] {Nicotiana
sylvestris}, partial (61%)
Length = 1374
Score = 78.2 bits (191), Expect = 9e-15
Identities = 51/184 (27%), Positives = 84/184 (44%), Gaps = 25/184 (13%)
Frame = +1
Query: 34 LYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVL 93
L+VG+L +V+ +QL ++F G V V V D +T RS G+G+V ++ + A L
Sbjct: 346 LFVGNLPFSVDSAQLAEIFENAGDVEMVEVIYDKSTGRSRGFGFVTMSSAAEVEAAAQQL 525
Query: 94 NFTPMNNKSIRVMYSHRDP------------------------SSRKSGTANIFIKNLDK 129
N ++ + +RV P G + + NL
Sbjct: 526 NGYVVDGRELRVNAGPPPPPRSENSRFGENPRFGGDRPRGPPRGGSSDGDNRVHVGNLAW 705
Query: 130 TIDHKALHDTFSSFGQIMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDK 188
+D+ AL F GQ++ K+ D SG+S+G+GFV F + D +AI L+G +N +
Sbjct: 706 GVDNLALESLFGEQGQVLEAKVIYDRESGRSRGFGFVTFSSADEVDSAIRTLDGADLNGR 885
Query: 189 QVFV 192
+ V
Sbjct: 886 AIRV 897
Score = 56.6 bits (135), Expect = 3e-08
Identities = 56/221 (25%), Positives = 93/221 (41%), Gaps = 11/221 (4%)
Frame = +1
Query: 200 DRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNF 258
D D+ S + ++V NL S L F G + ++ D + GRS+ FGFV
Sbjct: 307 DGDDTPSYSPNQRLFVGNLPFSVDSAQLAEIFENAGDVEMVEVIYDKSTGRSRGFGFVTM 486
Query: 259 ENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGL--- 315
+A + A + LNG VD E V R + RF + + D+ +G
Sbjct: 487 SSAAEVEAAAQQLNGYVVDGRELRVNAGPPPPPRSEN--SRFGENPRFGG-DRPRGPPRG 657
Query: 316 -------NLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRG 368
+++ NL + + L+ +F E G + KV I +G SRG
Sbjct: 658 GSSDGDNRVHVGNLAWGVDNLALESLFGEQGQVLEAKV-----------IYDRESGRSRG 804
Query: 369 SGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRR 409
GFV FS+ +E A+ ++G + + + V+ A + R+
Sbjct: 805 FGFVTFSSADEVDSAIRTLDGADLNGRAIRVSPADSRPKRQ 927
Score = 48.9 bits (115), Expect = 6e-06
Identities = 28/97 (28%), Positives = 52/97 (52%)
Frame = +1
Query: 34 LYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVL 93
++VG+L V++ L LF + GQV+ +V D + RS G+G+V F++ + A+ L
Sbjct: 682 VHVGNLAWGVDNLALESLFGEQGQVLEAKVIYDRESGRSRGFGFVTFSSADEVDSAIRTL 861
Query: 94 NFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKT 130
+ +N ++IRV + P + G I + ++T
Sbjct: 862 DGADLNGRAIRVSPADSRPKRQF*GFCLSSIPDQERT 972
>TC87747 weakly similar to PIR|C84768|C84768 hypothetical protein At2g35410
[imported] - Arabidopsis thaliana, partial (53%)
Length = 1177
Score = 76.6 bits (187), Expect = 3e-14
Identities = 58/197 (29%), Positives = 92/197 (46%), Gaps = 3/197 (1%)
Frame = +1
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEAL 271
N+ V NL S ++ D+K+ FG GT+ +++ DG+ K + FV ++ E A AV+
Sbjct: 304 NLIVFNLPWSLSKPDIKDLFGQCGTVIDVEIIKSKDGKGKGYTFVTMDSGEGAQAAVDKF 483
Query: 272 NGKKVDDEEWYV--GKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDE 329
N ++ V K KK T KE+ +Y NL
Sbjct: 484 NATEISGRILRVEFAKGFKKPRPPPPAP-----TPKEA------RYVIYASNLAWKARST 630
Query: 330 KLKEMFSE-FGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMN 388
L+++F+E F T S RV+ + P G S G GFV++ T EEA A+ +
Sbjct: 631 HLRDIFTENFKTPVS-----------ARVVFQVPGGKSAGYGFVSYHTEEEAEAAISALQ 777
Query: 389 GKMIVSKPLYVAVAQRK 405
GK ++ +PL V +++RK
Sbjct: 778 GKELLGRPLLVKISERK 828
Score = 62.8 bits (151), Expect = 4e-10
Identities = 54/215 (25%), Positives = 94/215 (43%), Gaps = 6/215 (2%)
Frame = +1
Query: 121 NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKL 180
N+ + NL ++ + D F G ++ +I G+ KGY FV ++ + AQ A+DK
Sbjct: 304 NLIVFNLPWSLSKPDIKDLFGQCGTVIDVEIIKSKDGKGKGYTFVTMDSGEGAQAAVDKF 483
Query: 181 NGMLINDKQVFV----GHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEF--GAY 234
N I+ + + V G + K +Y NL+ L++ F
Sbjct: 484 NATEISGRILRVEFAKGFKKPRPPPPAPTPKEARYVIYASNLAWKARSTHLRDIFTENFK 663
Query: 235 GTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQ 294
+++ V+ + G+S +GFV++ E+A A+ AL GK++ V K SER+
Sbjct: 664 TPVSARVVFQVPGGKSAGYGFVSYHTEEEAEAAISALQGKELLGRPLLV----KISERKV 831
Query: 295 ELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDE 329
+ G E K V G + +DD+ +E
Sbjct: 832 KEAGSEEVEEKVDDVQPEAGSEEVEEKVDDAQPEE 936
>TC92312 weakly similar to GP|4567275|gb|AAD23688.1| expressed protein
{Arabidopsis thaliana}, partial (29%)
Length = 1319
Score = 75.1 bits (183), Expect = 8e-14
Identities = 81/331 (24%), Positives = 135/331 (40%), Gaps = 39/331 (11%)
Frame = +1
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
+ ++NL + D FSS G + I +G SKG+ FV+F + A+NAI KL
Sbjct: 280 LIVRNLPFKAKENEIRDAFSSAGTVWEVFIPQKSDTGLSKGFAFVKFTCKQDAENAIRKL 459
Query: 181 NGMLINDKQVFVGHFLRKQ----DRDNVLSKTKFNNVYVKNLSESFTEDDLKN-----EF 231
NG + + V + K+ D ++ + + + TEDDL+N +
Sbjct: 460 NGSKFGSRLIAVDWAVPKKIFSSDTNDAPASEEGQQKVTDEDGSTTTEDDLENTDKKSDQ 639
Query: 232 GAYGTITSAV---------LMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWY 282
G I S V ++AD K + +A+D E++N V EE
Sbjct: 640 GDDSDIDSVVEEDVPSEDDFDKEADIARKVLNNLITSSAKD-----ESVNNDSVSSEE-- 798
Query: 283 VGKAQKKSEREQELKGRFEQTVKESVVDKFQGLN---------------LYLKNLDDSIT 327
+ K + ++ +KG +T KES DK ++ +++ NL +
Sbjct: 799 ----KNKPKSKETVKGADSKTSKES--DKVSDISKPETSKETEDDLHRTVFITNLPFELD 960
Query: 328 DEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEM 387
E+LK+ FS FG + + ++ RG+GF+ F T E A A+
Sbjct: 961 TEELKQRFSAFGEVEYF-----------APVLHQVTKRPRGTGFLKFKTAEAADNAISTA 1107
Query: 388 N-----GKMIVSKPLYVAVAQRKEDRRARLQ 413
N G ++ +PL V A K+ + Q
Sbjct: 1108NTASGMGILVKGRPLKVLKALDKKSAHDKEQ 1200
>BE998254 weakly similar to GP|14029147|gb| polyadenylated mRNA-binding
protein 2 {Anemia phyllitidis}, partial (12%)
Length = 211
Score = 74.3 bits (181), Expect = 1e-13
Identities = 35/69 (50%), Positives = 52/69 (74%), Gaps = 1/69 (1%)
Frame = +3
Query: 218 LSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKV- 276
L E+ T++DLK FG +G ITSA +M+DA+G+S+CFGFVNF++ +A AVE +NG +
Sbjct: 3 LYEASTDEDLKQLFGPFGMITSATVMKDANGKSRCFGFVNFQSPYSSAAAVEKVNGTAIN 182
Query: 277 DDEEWYVGK 285
DD+ +VG+
Sbjct: 183 DDKVLFVGR 209
Score = 50.1 bits (118), Expect = 3e-06
Identities = 25/59 (42%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Frame = +3
Query: 136 LHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLIN-DKQVFVG 193
L F FG I S + D +G+S+ +GFV F++ S+ A++K+NG IN DK +FVG
Sbjct: 30 LKQLFGPFGMITSATVMKDANGKSRCFGFVNFQSPYSSAAAVEKVNGTAINDDKVLFVG 206
Score = 47.0 bits (110), Expect = 2e-05
Identities = 27/71 (38%), Positives = 39/71 (54%)
Frame = +3
Query: 322 LDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEAS 381
L ++ TDE LK++F FG ITS +M+D NG SR GFV F +P ++
Sbjct: 3 LYEASTDEDLKQLFGPFGMITS------------ATVMKDANGKSRCFGFVNFQSPYSSA 146
Query: 382 RALGEMNGKMI 392
A+ ++NG I
Sbjct: 147 AAVEKVNGTAI 179
>BG644387 homologue to GP|6996560|emb oligouridylate binding protein
{Nicotiana plumbaginifolia}, partial (53%)
Length = 717
Score = 72.8 bits (177), Expect = 4e-13
Identities = 50/182 (27%), Positives = 94/182 (51%), Gaps = 9/182 (4%)
Frame = +2
Query: 10 TPAPVPAPS-----NGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVC 64
TPA + P +G +P + S+YVG++ V + L ++F+ G + ++
Sbjct: 38 TPALLAPPQIEPILSGNLPPGFDSSTCRSVYVGNIHPQVTEPLLQEVFSSTGPLEGCKLI 217
Query: 65 RDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTA---N 121
+ + YG+V++ + + AA A+ LN + + I+V +++ S+++ T+ N
Sbjct: 218 K----KEKSSYGFVDYFDRRSAALAIVTLNGRNLFGQPIKVNWAYT--SAQREDTSSHFN 379
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
IF+ +L + L+ FS + K+ D SG+S+G+GFV F + AQ+AI+ L
Sbjct: 380 IFVGDLSPEVTDATLYACFSVYPSCSDAKVMWDQKSGRSRGFGFVSFRNQQEAQSAINDL 559
Query: 181 NG 182
NG
Sbjct: 560 NG 565
Score = 72.0 bits (175), Expect = 7e-13
Identities = 49/170 (28%), Positives = 80/170 (46%), Gaps = 1/170 (0%)
Frame = +2
Query: 105 VMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGF 164
++ + P S ++++ N+ + L + FSS G + CK+ + YGF
Sbjct: 74 ILSGNLPPGFDSSTCRSVYVGNIHPQVTEPLLQEVFSSTGPLEGCKLIKK---EKSSYGF 244
Query: 165 VQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTE 224
V + SA AI LNG + + + V R++ S N++V +LS T+
Sbjct: 245 VDYFDRRSAALAIVTLNGRNLFGQPIKVNWAYTSAQREDTSSHF---NIFVGDLSPEVTD 415
Query: 225 DDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALNG 273
L F Y + + A +M D GRS+ FGFV+F N ++A A+ LNG
Sbjct: 416 ATLYACFSVYPSCSDAKVMWDQKSGRSRGFGFVSFRNQQEAQSAINDLNG 565
Score = 63.2 bits (152), Expect = 3e-10
Identities = 48/178 (26%), Positives = 79/178 (43%)
Frame = +2
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEAL 271
+VYV N+ TE L+ F + G + L++ +GFV++ + AA A+ L
Sbjct: 122 SVYVGNIHPQVTEPLLQEVFSSTGPLEGCKLIKK---EKSSYGFVDYFDRRSAALAIVTL 292
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKL 331
NG+ + + V A ++RE D N+++ +L +TD L
Sbjct: 293 NGRNLFGQPIKVNWAYTSAQRE----------------DTSSHFNIFVGDLSPEVTDATL 424
Query: 332 KEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNG 389
FS Y C+ D +V+ +G SRG GFV+F +EA A+ ++NG
Sbjct: 425 YACFSV------YPSCS-----DAKVMWDQKSGRSRGFGFVSFRNQQEAQSAINDLNG 565
Score = 30.8 bits (68), Expect = 1.7
Identities = 23/124 (18%), Positives = 53/124 (42%), Gaps = 3/124 (2%)
Frame = +2
Query: 316 NLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFS 375
++Y+ N+ +T+ L+E+FS G + K+ I ++ + GFV +
Sbjct: 122 SVYVGNIHPQVTEPLLQEVFSSTGPLEGCKL-----------IKKEKSSY----GFVDYF 256
Query: 376 TPEEASRALGEMNGKMIVSKPL---YVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRM 432
A+ A+ +NG+ + +P+ + + ++ED + + P ++
Sbjct: 257 DRRSAALAIVTLNGRNLFGQPIKVNWAYTSAQREDTSSHFNIFVGDLSPEVTDATLYACF 436
Query: 433 PLYP 436
+YP
Sbjct: 437 SVYP 448
>TC78142 similar to GP|7578881|gb|AAF64167.1| plastid-specific ribosomal
protein 2 precursor {Spinacia oleracea}, partial (65%)
Length = 819
Score = 72.4 bits (176), Expect = 5e-13
Identities = 61/211 (28%), Positives = 93/211 (43%), Gaps = 2/211 (0%)
Frame = +2
Query: 191 FVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGR 249
F HF+ ++ V S +YV N+ + + D+L+ +G + A +M D R
Sbjct: 197 FRKHFVVSSEQATVDSPA-LRKLYVGNIPRTVSNDELEKIVQEHGAVEKAEVMYDKYSKR 373
Query: 250 SKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVV 309
S+ F FV + EDA A E LNG ++ E V +K E G Q + + V
Sbjct: 374 SRRFAFVTMKTVEDANAAAEKLNGTEIGGREIKVNITEKPLTTE----GLPVQAGESTFV 541
Query: 310 DKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDP-NGVSRG 368
D +Y+ NL ++T + LK+ FSE G S KV R P S G
Sbjct: 542 D--SPYKVYVGNLAKNVTSDSLKKFFSEKGNALSAKV------------SRAPGTSKSSG 679
Query: 369 SGFVAFSTPEEASRALGEMNGKMIVSKPLYV 399
GFV FS+ E+ A+ N ++ + + V
Sbjct: 680 FGFVTFSSDEDVEAAISSFNNALLEGQKIRV 772
Score = 49.7 bits (117), Expect = 4e-06
Identities = 24/72 (33%), Positives = 39/72 (53%)
Frame = +2
Query: 34 LYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVL 93
+YVG+L NV L F++ G +S +V R T +S G+G+V F++ +D A+
Sbjct: 557 VYVGNLAKNVTSDSLKKFFSEKGNALSAKVSRAPGTSKSSGFGFVTFSSDEDVEAAISSF 736
Query: 94 NFTPMNNKSIRV 105
N + + IRV
Sbjct: 737 NNALLEGQKIRV 772
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,775,395
Number of Sequences: 36976
Number of extensions: 212580
Number of successful extensions: 2046
Number of sequences better than 10.0: 221
Number of HSP's better than 10.0 without gapping: 1467
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1865
length of query: 659
length of database: 9,014,727
effective HSP length: 102
effective length of query: 557
effective length of database: 5,243,175
effective search space: 2920448475
effective search space used: 2920448475
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148486.10


