

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148485.6 + phase: 0
(380 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
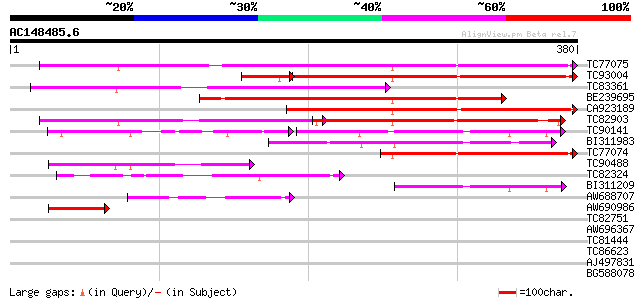
Score E
Sequences producing significant alignments: (bits) Value
TC77075 similar to GP|21593294|gb|AAM65243.1 putative carboxyl-t... 315 2e-86
TC93004 similar to GP|21537066|gb|AAM61407.1 unknown {Arabidopsi... 189 1e-53
TC83361 similar to PIR|E96729|E96729 unknown protein F5A18.27 [i... 196 1e-50
BE239695 homologue to GP|15529145|gb AT5g50150/MPF21_17 {Arabido... 193 1e-49
CA923189 weakly similar to GP|21389708|gb putative carboxyl-term... 178 3e-45
TC82903 similar to GP|21593294|gb|AAM65243.1 putative carboxyl-t... 151 5e-37
TC90141 similar to PIR|E84671|E84671 hypothetical protein At2g27... 100 1e-31
BI311983 similar to PIR|E84671|E846 hypothetical protein At2g273... 114 6e-26
TC77074 similar to GP|21593294|gb|AAM65243.1 putative carboxyl-t... 113 1e-25
TC90488 similar to PIR|E96729|E96729 unknown protein F5A18.27 [i... 98 6e-21
TC82324 similar to GP|15528706|dbj|BAB64772. hypothetical protei... 85 4e-17
BI311209 67 1e-11
AW688707 weakly similar to GP|21537066|gb| unknown {Arabidopsis ... 66 3e-11
AW690986 similar to GP|21389708|gb| putative carboxyl-terminal p... 54 8e-08
TC82751 similar to GP|3128171|gb|AAC16075.1| unknown protein {Ar... 30 0.006
AW696367 similar to GP|17065340|g putative Na+/H+ antiporter {Ar... 30 2.0
TC81444 PIR|C89872|C89872 hypothetical protein [imported] - Stap... 29 2.6
TC86623 similar to SP|O64765|UAP1_ARATH Probable UDP-N-acetylglu... 28 5.8
AJ497831 homologue to GP|3608175|dbj proliferating cell nuclear ... 28 5.8
BG588078 similar to GP|4314359|gb| unknown protein {Arabidopsis ... 28 5.8
>TC77075 similar to GP|21593294|gb|AAM65243.1 putative carboxyl-terminal
peptidase {Arabidopsis thaliana}, partial (76%)
Length = 2015
Score = 315 bits (806), Expect = 2e-86
Identities = 159/398 (39%), Positives = 241/398 (59%), Gaps = 38/398 (9%)
Frame = +1
Query: 21 SLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTI--------- 71
S+ + E++ LK NRP VK+IKS +GDIIDCV + QPAFDHP LK+H I
Sbjct: 370 SIQKLEVKKHLKNLNRPPVKSIKSPDGDIIDCVHVSHQPAFDHPELKDHKIQMRPNFHPE 549
Query: 72 ---------------------QTWHKSGRCPKGTVPIRRIQKQDLLRAATLDRFGLKQSS 110
Q W K+G C +GT+PIRR + D+LRA+++ FG K+
Sbjct: 550 RKTFGESKVSSNSNSNSKPITQLWQKNGMCSEGTIPIRRTRTNDILRASSVQNFGKKKQK 729
Query: 111 SFVNSKNT-TISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDS 169
S K + + SG + ++ G +F GA+A INVW+PK+ P++
Sbjct: 730 STPQPKPAKPLPDILTQSGHQHAIA--------YVEGGDFYGAKATINVWDPKIQQPNEF 885
Query: 170 TTAQIWLKAGN-GNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFV 228
+ +QIW+ AG G + SIEAGW V+P LYG+++TRLF+YWT+D+Y++TGC++L CSGF+
Sbjct: 886 SLSQIWILAGAFGQDLNSIEAGWQVSPDLYGDNNTRLFTYWTSDAYQATGCYNLLCSGFI 1065
Query: 229 QTSNTVALGGGINPISSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLK 283
Q +N +ALG I+P+S+ +QY+++I + G+WW++ +D +GYWP LF+ L
Sbjct: 1066QINNGIALGASISPLSNYGSSQYDISILVWKDPKEGNWWMQFGNDHVLGYWPAPLFSYLT 1245
Query: 284 HSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPES 343
SA++++WGG+V +S+ HT TQMGSGH DE +G A Y +NI++ D L+ P+
Sbjct: 1246ESASMIEWGGEVVNSE-SDGQHTSTQMGSGHFPDEGFGKASYFKNIQVVDGDNKLRAPKD 1422
Query: 344 INVASQEPNCYSAFNDEDVQEPT-FYFGGPGQSSPSCP 380
+ +++ NCY+ T FY+GGPG+ +P+CP
Sbjct: 1423LGTYTEKDNCYNVKTGNAGDWGTYFYYGGPGR-NPNCP 1533
>TC93004 similar to GP|21537066|gb|AAM61407.1 unknown {Arabidopsis
thaliana}, partial (53%)
Length = 827
Score = 189 bits (480), Expect(2) = 1e-53
Identities = 89/197 (45%), Positives = 131/197 (66%), Gaps = 6/197 (3%)
Frame = +1
Query: 190 GWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGT 249
G V+P LYG++ RLF+YWT+DSY++TGC++L C+GF+QT++ +A+G I+P+SS +G
Sbjct: 118 GSQVSPELYGDNRPRLFTYWTSDSYQATGCYNLLCAGFIQTNSKIAIGAAISPVSSYTGN 297
Query: 250 QYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSP 304
QY+++I VG+WW+ + +GYWP ELFT L AT+V+WGG+V +S+
Sbjct: 298 QYDISILIWKDPRVGNWWMSFGDNTLVGYWPAELFTHLADHATMVEWGGEVVNSRT-DGR 474
Query: 305 HTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFNDEDVQE 364
HT TQMGSGH A++ +G A Y RN+ I D L +SI+ ++ NCY N +
Sbjct: 475 HTSTQMGSGHFAEDGFGKASYFRNLEIVDIDNTLSSVQSISTLAENTNCYDIQNSYSNEW 654
Query: 365 PT-FYFGGPGQSSPSCP 380
T FY+GGPG ++P CP
Sbjct: 655 GTYFYYGGPG-NNPKCP 702
Score = 38.5 bits (88), Expect(2) = 1e-53
Identities = 15/38 (39%), Positives = 28/38 (73%), Gaps = 2/38 (5%)
Frame = +3
Query: 156 INVWNPKVDLPDDSTTAQIWLKAG--NGNEFESIEAGW 191
I+VW+P +++ ++ + +QIW+ +G +G + SIEAGW
Sbjct: 6 ISVWDPSIEVMNEFSLSQIWVLSGSFDGPDLNSIEAGW 119
>TC83361 similar to PIR|E96729|E96729 unknown protein F5A18.27 [imported] -
Arabidopsis thaliana, partial (57%)
Length = 1095
Score = 196 bits (498), Expect = 1e-50
Identities = 106/264 (40%), Positives = 153/264 (57%), Gaps = 23/264 (8%)
Frame = +2
Query: 15 FVAGAISLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHT---- 70
+ AG+ L I L N+PAVKTI+S +GD+IDC+ + QPAFDHP LK H
Sbjct: 329 YKAGSELLKLRRIRTHLMKINKPAVKTIQSPDGDLIDCILSHHQPAFDHPKLKGHKPLDP 508
Query: 71 -----------------IQTWHKSGR-CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSF 112
+Q W SG CP+GTVPIRR + D+LRA+++ RFG K
Sbjct: 509 PERPNGYYNNGEKVSERLQLWTDSGEECPEGTVPIRRTTEDDILRASSIKRFGRKPKPVR 688
Query: 113 VNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTA 172
+S S DH L +G+ + GA+A+INVW P V + + +
Sbjct: 689 RDS-----------------TSSDHEHAILFVNGNQYFGAKANINVWAPHVTDGYEFSLS 817
Query: 173 QIWLKAGN-GNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTS 231
QIW+ AG+ GN+ ++EAGW V+P LYG++ R F+YWTTD+Y++TGC++L CSGF+QT+
Sbjct: 818 QIWVIAGSFGNDLNTLEAGWQVSPELYGDNYPRFFTYWTTDAYQTTGCYNLLCSGFIQTN 997
Query: 232 NTVALGGGINPISSDSGTQYELNI 255
N +A+G I P S+ + Q+++ I
Sbjct: 998 NKIAIGATIXPTSAYNSGQFDIGI 1069
>BE239695 homologue to GP|15529145|gb AT5g50150/MPF21_17 {Arabidopsis
thaliana}, partial (50%)
Length = 637
Score = 193 bits (490), Expect = 1e-49
Identities = 90/212 (42%), Positives = 142/212 (66%), Gaps = 6/212 (2%)
Frame = +2
Query: 128 GSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGN-GNEFES 186
G S +H+ V + +G + GA+A INVW P+V + + +QIW+ AG+ GN+ +
Sbjct: 2 GDSTGNGHEHAVVFV--NGEQYYGAKASINVWTPRVTDQYEFSLSQIWVIAGSFGNDLNT 175
Query: 187 IEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISSD 246
+EAGW V+P LYG++ R F+YWTTD+Y++TGC++L CSGF+QT+N +A+G I+P S
Sbjct: 176 LEAGWQVSPELYGDNYPRFFTYWTTDAYQATGCYNLLCSGFIQTNNRIAIGAAISPRSFY 355
Query: 247 SGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVK 301
SG Q+++ + + GHWWL+ + +GYWP LF+ L+ A++VQ+GG++ +S+
Sbjct: 356 SGRQFDIGLMIWKDPKHGHWWLEFGSGLLVGYWPANLFSHLRSHASMVQFGGEIVNSR-S 532
Query: 302 KSPHTKTQMGSGHLADEKYGHACYMRNIRIKD 333
+ HT TQMGSGH + E + A Y RN+++ D
Sbjct: 533 RGYHTGTQMGSGHFSGEGFRKAAYFRNLQVVD 628
>CA923189 weakly similar to GP|21389708|gb putative carboxyl-terminal
proteinase {Gossypium hirsutum}, partial (39%)
Length = 778
Score = 178 bits (452), Expect = 3e-45
Identities = 87/200 (43%), Positives = 126/200 (62%), Gaps = 5/200 (2%)
Frame = -2
Query: 186 SIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISS 245
+IEAGW V+ +YG++ R+F YWT + YK TGC++L C GFVQTS T ALGG ++P S+
Sbjct: 774 TIEAGWQVSHQIYGDYLPRIFVYWTANGYKGTGCYNLRCPGFVQTSKTFALGGALSPPST 595
Query: 246 DSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQV 300
+G Q E+ + + G+WWL+ IGYWP LFTSLK SAT V++GG+V+
Sbjct: 594 YNGRQIEITLLIYKDQKTGNWWLEYGSGNIIGYWPSSLFTSLKDSATTVEFGGEVYIKST 415
Query: 301 KKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFNDE 360
H+ TQMGSGH ADE A Y R++++ ++ L ++NV ++EP CY+
Sbjct: 414 --GTHSSTQMGSGHFADEGSSKASYFRDMQVVNSDNKLIPLSNLNVFAEEPKCYNIIKGT 241
Query: 361 DVQEPTFYFGGPGQSSPSCP 380
+ FY+GGPG++ +CP
Sbjct: 240 SNEGNYFYYGGPGKNM-NCP 184
>TC82903 similar to GP|21593294|gb|AAM65243.1 putative carboxyl-terminal
peptidase {Arabidopsis thaliana}, partial (88%)
Length = 1155
Score = 151 bits (381), Expect = 5e-37
Identities = 77/182 (42%), Positives = 112/182 (61%), Gaps = 13/182 (7%)
Frame = +3
Query: 204 RLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELNI-----DEV 258
RLF+YWT+D+Y++TGC +L CSGF+Q S+ +A+G I+PISS + +++I +
Sbjct: 630 RLFTYWTSDAYQATGCNNLLCSGFIQVSSDIAMGASISPISSYRXSXXDISILIWKDPKE 809
Query: 259 GHWWLK-ENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLAD 317
GHWW++ N +GYWP LF+ L SAT+++WGG+V +S HT T MGSGH +
Sbjct: 810 GHWWMQFGNQGTVLGYWPSFLFSYLPDSATMIEWGGEVVNS-XPDGQHTSTXMGSGHFPE 986
Query: 318 EKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFNDEDVQEPT-------FYFG 370
E +G A Y RNI+ D+S LK P+ + ++ PNCY DVQ + FY+
Sbjct: 987 EGFGKASYFRNIQXVDSSNNLKAPKXLGTYTEHPNCY------DVQTGSNGDWGHFFYYX 1148
Query: 371 GP 372
GP
Sbjct: 1149GP 1154
Score = 149 bits (375), Expect = 2e-36
Identities = 84/233 (36%), Positives = 123/233 (52%), Gaps = 40/233 (17%)
Frame = +2
Query: 21 SLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTI--------- 71
S + E+ L N+P VKTI+S +GDIIDCV + KQPAFDHP LK+H I
Sbjct: 17 SRQKFEVNKHLNRLNKPPVKTIQSPDGDIIDCVPVSKQPAFDHPFLKDHKIQMRPNFHPE 196
Query: 72 --------------------QTWHKSGRCPKGTVPIRRIQKQDLLRAATLDRFGLKQSSS 111
Q WH +G+C +GT+PIRR +++D+LRA++ R+G K+ S
Sbjct: 197 GLFEENKLDDNKEKSSTPINQLWHANGKCSEGTIPIRRTKEEDVLRASSAKRYGRKKHKS 376
Query: 112 FVNSKNTTISNFSKLSGSSNVVSED-HSGVHLATSGSNFIGAEADINVWNPKVDLPDDST 170
F + S ++V++ H G F GA+A INVW PK+ ++ +
Sbjct: 377 FAKPR----------SAEPDLVNQSGHQHAIAYVEGDKFYGAKATINVWEPKIQQTNEFS 526
Query: 171 TAQIWLKAGN-GNEFESIEAGWMVNPGLYGNHDTR---------LFSYWTTDS 213
+QIW+ G+ G + SIEAGW V+P LYG+++T+ + SYW S
Sbjct: 527 LSQIWVLRGSFGQDLNSIEAGWQVSPDLYGDNNTQAIHLLDE*CISSYWLQQS 685
>TC90141 similar to PIR|E84671|E84671 hypothetical protein At2g27320
[imported] - Arabidopsis thaliana, partial (18%)
Length = 1166
Score = 100 bits (250), Expect(2) = 1e-31
Identities = 64/189 (33%), Positives = 94/189 (48%), Gaps = 9/189 (4%)
Frame = +1
Query: 193 VNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNT--VALGGGINPISSDSGTQ 250
V P LY ++ L S WT D +K TGC++ C GFVQ ++ +LG I+P +S T+
Sbjct: 547 VFPSLYHDNQLLLTSRWTADGFKQTGCYNDNCPGFVQVNSNKDYSLGIVISPTNSIGPTE 726
Query: 251 YELNIDEVGHWW-LKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQ 309
+ + GHWW L + I +GYWP ELF L A+ +++GGQ ++ SP
Sbjct: 727 KDRS---TGHWWLLMDPKSIQVGYWPRELFNHLGMGASKIRFGGQTYAPPNTNSP----P 885
Query: 310 MGSGHLADEKYGHACYMRNIRIKD---NSLMLKYPESINVASQEPNCYSAFN---DEDVQ 363
MGSG L EK+ ++ +M +RI D N +K NCY + +
Sbjct: 886 MGSGRLPKEKFENSGFMGQLRIIDSQYNEADVKPENMKPYRDTNSNCYDVIYNGFEGRLH 1065
Query: 364 EPTFYFGGP 372
F +GGP
Sbjct: 1066RQAFLYGGP 1092
Score = 53.5 bits (127), Expect(2) = 1e-31
Identities = 53/189 (28%), Positives = 80/189 (42%), Gaps = 24/189 (12%)
Frame = +3
Query: 26 EIEAKLKL-----HNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQTWHKSGR- 79
++EA+ K+ +N +I+ D DCVDIYKQPA HP+LK H IQ + +
Sbjct: 39 KVEARSKILLDETNNFTLSYSIEDFVADKFDCVDIYKQPALQHPLLKKHKIQLFPTFAKN 218
Query: 80 --------------CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSK 125
CP G VPI R G + ++ +S I +F +
Sbjct: 219 IVRNRPSYGKTADDCPLGKVPIYN------------SRGGHQIITN--SSSKLQIDDFQR 356
Query: 126 LSGSSNVVSEDHSGVHLAT----SGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGNG 181
S S + G H T + F GA A I ++ V S + IW+++G+G
Sbjct: 357 HSKS-------NPGYHTVTLDTIQNTIFHGAYAGITGYDLSVQAKQYS-MSYIWVESGSG 512
Query: 182 NEFESIEAG 190
+ SI+ G
Sbjct: 513 TQLNSIKVG 539
>BI311983 similar to PIR|E84671|E846 hypothetical protein At2g27320
[imported] - Arabidopsis thaliana, partial (16%)
Length = 593
Score = 114 bits (285), Expect = 6e-26
Identities = 71/201 (35%), Positives = 103/201 (50%), Gaps = 8/201 (3%)
Frame = +1
Query: 174 IWLKAGNGNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNT 233
IW+ +G E I G+ V+PGLYG+ RL SYWT D K TGC++ C GFVQ ++
Sbjct: 10 IWIASGPPTELNIILTGFGVHPGLYGDSQLRLTSYWTVDG-KKTGCYNQLCPGFVQVNHD 186
Query: 234 V--ALGGGINPISS-DSGTQYELNID-----EVGHWWLKENHDIPIGYWPVELFTSLKHS 285
ALG ++P + S T+Y I HWWL + I +GYWP ELFT L
Sbjct: 187 KENALGSVLSPTTPIGSTTKYVAPIKIKQDRSTSHWWLIIHESIYVGYWPKELFTHLSKG 366
Query: 286 ATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESIN 345
A +++GGQ ++ SP MGSG L EK+ ++ M + I D+ ++
Sbjct: 367 AAFIRFGGQTYAPPNNDSP----PMGSGRLPKEKFPNSGLMGELEIIDSGY-----NEMD 519
Query: 346 VASQEPNCYSAFNDEDVQEPT 366
V ++ Y+ N + + PT
Sbjct: 520 VNPEDMKPYTDTNSKCMTGPT 582
>TC77074 similar to GP|21593294|gb|AAM65243.1 putative carboxyl-terminal
peptidase {Arabidopsis thaliana}, partial (32%)
Length = 1167
Score = 113 bits (282), Expect = 1e-25
Identities = 53/138 (38%), Positives = 86/138 (61%), Gaps = 6/138 (4%)
Frame = +1
Query: 249 TQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKS 303
+QY+++I + G+WW++ +D +GYWP LF+ L SA++++WGG+V +S+
Sbjct: 28 SQYDISILVWKDPKEGNWWMQFGNDHVLGYWPAPLFSYLTESASMIEWGGEVVNSE-SDG 204
Query: 304 PHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFNDEDVQ 363
HT TQMGSGH DE +G A Y +NI++ D L+ P+ + +++ NCY+
Sbjct: 205 QHTSTQMGSGHFPDEGFGKASYFKNIQVVDGDNKLRAPKDLGTYTEKDNCYNVKTGNAGD 384
Query: 364 EPT-FYFGGPGQSSPSCP 380
T FY+GGPG+ +P+CP
Sbjct: 385 WGTYFYYGGPGR-NPNCP 435
>TC90488 similar to PIR|E96729|E96729 unknown protein F5A18.27 [imported] -
Arabidopsis thaliana, partial (32%)
Length = 648
Score = 97.8 bits (242), Expect = 6e-21
Identities = 59/159 (37%), Positives = 86/159 (53%), Gaps = 21/159 (13%)
Frame = +1
Query: 27 IEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNH-----------TIQTWH 75
I ++LK N+PAVKTI+S +GDIIDCV +KQPAFDHP+LK QT
Sbjct: 148 ITSRLKQINKPAVKTIQSSDGDIIDCVVSHKQPAFDHPLLKGQKPLDPLERSKGNNQTDI 327
Query: 76 KSGR----------CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSK 125
SG+ CP+GT+PIRR +++D+LRA+++D FG K S
Sbjct: 328 LSGKFQLWSLYDESCPEGTIPIRRAKEEDILRASSIDTFGRKLSQV-------------- 465
Query: 126 LSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVD 164
+++ H + + +G + GA+A IN+W P V+
Sbjct: 466 ---ATDATEYKHVHSYGSVTGDRYYGAKATINLWAPHVE 573
>TC82324 similar to GP|15528706|dbj|BAB64772. hypothetical protein~similar
to Arabidopsis thaliana chromosome 5 AAF04872.1,
partial (16%)
Length = 701
Score = 85.1 bits (209), Expect = 4e-17
Identities = 62/197 (31%), Positives = 93/197 (46%), Gaps = 4/197 (2%)
Frame = +1
Query: 32 KLHNRPAVKTIKSENGDIIDCVDIYKQP-AFDHPVLKNHTIQTWHKSGRCPKGTVPIRRI 90
+ H VKTI+ D+ K+P + + + +Q W +GT IRR
Sbjct: 124 EFHKPAVVKTIQ----------DLPKRPIGHNQTDILSDNMQFWSS-----EGTHSIRRT 258
Query: 91 QKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGSNFI 150
KQD+LR ++ RFG K + N+ +H +G F
Sbjct: 259 -KQDILRTKSISRFGRKLNQVGTNTG-------------------EHLYAIARVNGDKFY 378
Query: 151 GAEADINVWNPKVDLP---DDSTTAQIWLKAGNGNEFESIEAGWMVNPGLYGNHDTRLFS 207
GA+A INVW P V+ D+ + +QIW+ AG+ E +IEAGW +P G++ RLF
Sbjct: 379 GAKATINVWRPYVENNSNNDEFSLSQIWVTAGSDGELNTIEAGWRADPKFNGDNLPRLFI 558
Query: 208 YWTTDSYKSTGCFDLTC 224
+ T D Y+ TGC++L C
Sbjct: 559 FMTKDGYQ-TGCYNLGC 606
>BI311209
Length = 536
Score = 67.0 bits (162), Expect = 1e-11
Identities = 41/122 (33%), Positives = 58/122 (46%), Gaps = 7/122 (5%)
Frame = +3
Query: 259 GHWWL-KENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLAD 317
GHWWL + I +GYWP ELF L A+ +++GGQ ++ SP MGSG L
Sbjct: 117 GHWWLLMDPKSIQVGYWPRELFNHLGMGASKIRFGGQTYAPPNTNSP----PMGSGRLPK 284
Query: 318 EKYGHACYMRNIRIKD---NSLMLKYPESINVASQEPNCYSAFN---DEDVQEPTFYFGG 371
EK+ ++ +M +RI D N +K NCY + + F +GG
Sbjct: 285 EKFENSGFMGQLRIIDSQYNEADVKPENMKPYRDTNSNCYDVIYNGFEGRLHRQAFLYGG 464
Query: 372 PG 373
PG
Sbjct: 465 PG 470
>AW688707 weakly similar to GP|21537066|gb| unknown {Arabidopsis thaliana},
partial (12%)
Length = 584
Score = 65.9 bits (159), Expect = 3e-11
Identities = 36/112 (32%), Positives = 62/112 (55%)
Frame = +3
Query: 80 CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSG 139
CP+G+VPIRR +++D+LRA++++ FG K +N + + + + V
Sbjct: 48 CPEGSVPIRRTKEEDILRASSINTFGRK-----LNQVGMDTTKYKHVHSTGYV------- 191
Query: 140 VHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGNEFESIEAGW 191
+G + GA+A IN+W+P V+ + + +QIWL G + +IEAGW
Sbjct: 192 -----TGDLYYGAKATINLWSPHVEGEKEFSLSQIWLTTGRNS--NTIEAGW 326
>AW690986 similar to GP|21389708|gb| putative carboxyl-terminal proteinase
{Gossypium hirsutum}, partial (14%)
Length = 621
Score = 54.3 bits (129), Expect = 8e-08
Identities = 25/41 (60%), Positives = 32/41 (77%)
Frame = +2
Query: 27 IEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLK 67
I A LK N+ VKTIKS +GD+IDC+ ++QPAFDHP+LK
Sbjct: 449 IRAHLKKINKLPVKTIKSPDGDLIDCIISHQQPAFDHPLLK 571
>TC82751 similar to GP|3128171|gb|AAC16075.1| unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 744
Score = 30.0 bits (66), Expect(2) = 0.006
Identities = 12/14 (85%), Positives = 12/14 (85%)
Frame = +3
Query: 58 QPAFDHPVLKNHTI 71
QPAFDHP LKNH I
Sbjct: 252 QPAFDHPDLKNHKI 293
Score = 26.9 bits (58), Expect(2) = 0.006
Identities = 13/41 (31%), Positives = 21/41 (50%), Gaps = 1/41 (2%)
Frame = +1
Query: 72 QTWHKSGRCPKGTVPIRRIQ-KQDLLRAATLDRFGLKQSSS 111
Q WHK G C + T+ ++ + D+LR ++ F K S
Sbjct: 385 QVWHKIGMCSEETILYQKYK**YDILRTGSIQNFFKKNQRS 507
>AW696367 similar to GP|17065340|g putative Na+/H+ antiporter {Arabidopsis
thaliana}, partial (15%)
Length = 656
Score = 29.6 bits (65), Expect = 2.0
Identities = 15/35 (42%), Positives = 22/35 (62%)
Frame = +2
Query: 334 NSLMLKYPESINVASQEPNCYSAFNDEDVQEPTFY 368
N LML ESI++AS EP C + +V+ P++Y
Sbjct: 35 NILMLSVEESIDLASSEPLCDWKGLEANVRFPSYY 139
>TC81444 PIR|C89872|C89872 hypothetical protein [imported] - Staphylococcus
aureus (strain N315), partial (16%)
Length = 1434
Score = 29.3 bits (64), Expect = 2.6
Identities = 14/29 (48%), Positives = 19/29 (65%)
Frame = -2
Query: 159 WNPKVDLPDDSTTAQIWLKAGNGNEFESI 187
WN ++ PDD T A+ + G+ NEFESI
Sbjct: 905 WNNHLEYPDDWTEAEYY---GDTNEFESI 828
>TC86623 similar to SP|O64765|UAP1_ARATH Probable UDP-N-acetylglucosamine
pyrophosphorylase (EC 2.7.7.23). [Mouse-ear cress],
partial (93%)
Length = 2200
Score = 28.1 bits (61), Expect = 5.8
Identities = 11/15 (73%), Positives = 12/15 (79%)
Frame = +2
Query: 204 RLFSYWTTDSYKSTG 218
R+F YWTT S KSTG
Sbjct: 632 RMFQYWTTSS*KSTG 676
>AJ497831 homologue to GP|3608175|dbj proliferating cell nuclear antigen
{Pisum sativum}, partial (59%)
Length = 533
Score = 28.1 bits (61), Expect = 5.8
Identities = 17/47 (36%), Positives = 24/47 (50%)
Frame = +2
Query: 128 GSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQI 174
G + V+S GV +T G IGA + N VD PDD+T ++
Sbjct: 167 GDTVVISVTKEGVKFSTKGD--IGAANIVCRQNTTVDKPDDATVIEM 301
>BG588078 similar to GP|4314359|gb| unknown protein {Arabidopsis thaliana},
partial (20%)
Length = 623
Score = 28.1 bits (61), Expect = 5.8
Identities = 17/61 (27%), Positives = 26/61 (41%), Gaps = 4/61 (6%)
Frame = +2
Query: 234 VALGGGINP---ISSDSGTQYEL-NIDEVGHWWLKENHDIPIGYWPVELFTSLKHSATLV 289
V L INP + D QY N D H W+ + D P+G+W + ++ +
Sbjct: 2 VLLTNPINPQFRLEVDDKYQYSWENRDNTVHGWIGFDSDPPVGFWMITPSNEFRNGGPIK 181
Query: 290 Q 290
Q
Sbjct: 182 Q 184
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,671,120
Number of Sequences: 36976
Number of extensions: 177704
Number of successful extensions: 909
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 870
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 873
length of query: 380
length of database: 9,014,727
effective HSP length: 98
effective length of query: 282
effective length of database: 5,391,079
effective search space: 1520284278
effective search space used: 1520284278
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148485.6


