

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148485.5 + phase: 0
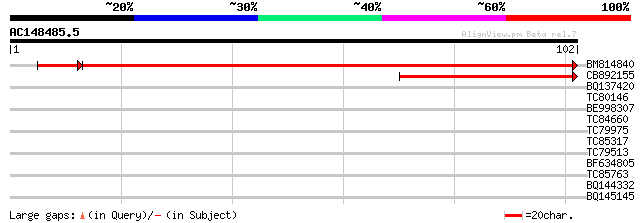
(102 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BM814840 weakly similar to GP|15128241|db helicase-like protein ... 169 2e-43
CB892155 similar to PIR|D86481|D86 hypothetical protein AAG28292... 54 7e-09
BQ137420 37 0.001
TC80146 36 0.003
BE998307 33 0.023
TC84660 similar to PIR|D86481|D86481 hypothetical protein AAG282... 30 0.15
TC79975 similar to GP|16417944|gb|AAL18924.1 acetyl Co-A acetylt... 25 3.7
TC85317 homologue to GP|6996529|emb|CAB75428.1 enolase {Lupinus ... 25 6.3
TC79513 weakly similar to GP|9885806|gb|AAG01535.1| stellacyanin... 25 6.3
BF634805 similar to GP|12321742|gb| peptide chain release factor... 24 8.2
TC85763 similar to GP|12083298|gb|AAG48808.1 unknown protein {Ar... 24 8.2
BQ144332 similar to GP|398168|emb|C keratin 2 epidermis {Mus mus... 24 8.2
BQ145145 similar to SP|O64407|AMYB_ Beta-amylase (EC 3.2.1.2) (1... 24 8.2
>BM814840 weakly similar to GP|15128241|db helicase-like protein {Oryza
sativa (japonica cultivar-group)}, partial (6%)
Length = 733
Score = 169 bits (428), Expect(2) = 2e-43
Identities = 85/89 (95%), Positives = 86/89 (96%)
Frame = +3
Query: 14 NGTRLIIVSLGKNVICARVIGGAHAGEVVYIPRMNLIPSGANVSITFERRQFPLVLSFAM 73
+GTRLIIVSLGKNVICARVIGG HAGEV YIPRMNLIPSGANVSITFER QFPLVLSFAM
Sbjct: 27 HGTRLIIVSLGKNVICARVIGGTHAGEVSYIPRMNLIPSGANVSITFERCQFPLVLSFAM 206
Query: 74 TINKSQGQTLTSVGLYLPRPVFTHGQLYV 102
TINKSQGQTLTSVGLYLPRPVFTHGQLYV
Sbjct: 207 TINKSQGQTLTSVGLYLPRPVFTHGQLYV 293
Score = 20.8 bits (42), Expect(2) = 2e-43
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = +1
Query: 6 VDQASGLC 13
VDQASGLC
Sbjct: 4 VDQASGLC 27
>CB892155 similar to PIR|D86481|D86 hypothetical protein AAG28292.1
[imported] - Arabidopsis thaliana, partial (4%)
Length = 572
Score = 54.3 bits (129), Expect = 7e-09
Identities = 24/32 (75%), Positives = 28/32 (87%)
Frame = -2
Query: 71 FAMTINKSQGQTLTSVGLYLPRPVFTHGQLYV 102
FAMTINKSQGQ+L +G+YLP VF+HGQLYV
Sbjct: 358 FAMTINKSQGQSLKHIGVYLPSSVFSHGQLYV 263
>BQ137420
Length = 690
Score = 37.0 bits (84), Expect = 0.001
Identities = 18/38 (47%), Positives = 26/38 (68%)
Frame = +2
Query: 42 VYIPRMNLIPSGANVSITFERRQFPLVLSFAMTINKSQ 79
+YI ++NLI SGANVS+ F+R + L F M +KS+
Sbjct: 503 IYILQINLILSGANVSVKFKRHRLLLGFGFTMVGSKSE 616
>TC80146
Length = 476
Score = 35.8 bits (81), Expect = 0.003
Identities = 14/33 (42%), Positives = 25/33 (75%)
Frame = -3
Query: 69 LSFAMTINKSQGQTLTSVGLYLPRPVFTHGQLY 101
+ + TINKS+ Q+L+ + +YL RP+F+H ++Y
Sbjct: 465 IPYFKTINKSR*QSLSYMKIYLSRPIFSHEEMY 367
>BE998307
Length = 624
Score = 32.7 bits (73), Expect = 0.023
Identities = 21/61 (34%), Positives = 33/61 (53%), Gaps = 1/61 (1%)
Frame = +2
Query: 42 VYIPRMNLIPSGANVSITFERRQFPLVLSFAMTINKSQGQTLTSVGLYL-PRPVFTHGQL 100
+YI ++NLI S ANVS+ F+R + L + K + + + Y+ R VFT QL
Sbjct: 386 IYILQINLILSRANVSVKFKRHRLLLCFVLQWLLVKVKDKFYERLVYYIKTRSVFTDDQL 565
Query: 101 Y 101
+
Sbjct: 566 H 568
>TC84660 similar to PIR|D86481|D86481 hypothetical protein AAG28292.1
[imported] - Arabidopsis thaliana, partial (1%)
Length = 1009
Score = 30.0 bits (66), Expect = 0.15
Identities = 11/21 (52%), Positives = 17/21 (80%)
Frame = +2
Query: 82 TLTSVGLYLPRPVFTHGQLYV 102
++ + G+YLP+P+F HG LYV
Sbjct: 68 SMATKGMYLPQPIF*HG*LYV 130
>TC79975 similar to GP|16417944|gb|AAL18924.1 acetyl Co-A acetyltransferase
{Hevea brasiliensis}, partial (73%)
Length = 1019
Score = 25.4 bits (54), Expect = 3.7
Identities = 10/33 (30%), Positives = 19/33 (57%)
Frame = -2
Query: 14 NGTRLIIVSLGKNVICARVIGGAHAGEVVYIPR 46
NG R++++S +IC +HA ++ IP+
Sbjct: 619 NGIRVLLISSYGMLICTHFCTNSHAKVIIDIPK 521
>TC85317 homologue to GP|6996529|emb|CAB75428.1 enolase {Lupinus luteus},
complete
Length = 1755
Score = 24.6 bits (52), Expect = 6.3
Identities = 14/54 (25%), Positives = 23/54 (41%)
Frame = +1
Query: 34 GGAHAGEVVYIPRMNLIPSGANVSITFERRQFPLVLSFAMTINKSQGQTLTSVG 87
GG+HAG + + ++P GA+ + + I K GQ +VG
Sbjct: 598 GGSHAGNKLAMQEFMVLPVGASSFKEAMKMGVEVYHHLKAVIKKKYGQDAVNVG 759
>TC79513 weakly similar to GP|9885806|gb|AAG01535.1| stellacyanin-like
protein CASLP1 precursor {Capsicum annuum}, partial
(33%)
Length = 1093
Score = 24.6 bits (52), Expect = 6.3
Identities = 22/74 (29%), Positives = 34/74 (45%)
Frame = -3
Query: 16 TRLIIVSLGKNVICARVIGGAHAGEVVYIPRMNLIPSGANVSITFERRQFPLVLSFAMTI 75
T +++V +GK V RV+G + +PR+ S N ER +F L S +
Sbjct: 266 TNVLLVFIGKKVQDLRVVGV*TEPSI*CVPRLWTKKSY*N*DTLIERLKFYLFNSDTRSK 87
Query: 76 NKSQGQTLTSVGLY 89
+KS +L LY
Sbjct: 86 DKSISFSLH*QSLY 45
>BF634805 similar to GP|12321742|gb| peptide chain release factor 2 putative
{Arabidopsis thaliana}, partial (18%)
Length = 657
Score = 24.3 bits (51), Expect = 8.2
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 3/38 (7%)
Frame = +1
Query: 58 ITFERRQFPLV---LSFAMTINKSQGQTLTSVGLYLPR 92
I +R FP+ + F ++ + +TLT G YLPR
Sbjct: 208 IRLHQRCFPVATYHMPFLSSLPRGFQETLTLTGYYLPR 321
>TC85763 similar to GP|12083298|gb|AAG48808.1 unknown protein {Arabidopsis
thaliana}, partial (93%)
Length = 1207
Score = 24.3 bits (51), Expect = 8.2
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = -2
Query: 16 TRLIIVSLGKNVICARVIGGAHAGEV 41
TR+ I SLG+ +C RV+ + G +
Sbjct: 240 TRIRIGSLGRCTVCLRVLLSSRCGRI 163
>BQ144332 similar to GP|398168|emb|C keratin 2 epidermis {Mus musculus},
partial (7%)
Length = 1163
Score = 24.3 bits (51), Expect = 8.2
Identities = 16/64 (25%), Positives = 32/64 (50%), Gaps = 7/64 (10%)
Frame = -3
Query: 6 VDQASGLCNGTRLIIVSLGKNVICARVIGGAHAGEV-------VYIPRMNLIPSGANVSI 58
+++A C TRL S K+ +CA+ GA A V +++ + L+P+ + + +
Sbjct: 1146 IEEAGDCCISTRLRCRS*KKHPLCAQ---GASAATV*SPHTR*IFLRKFGLLPAPSTLLV 976
Query: 59 TFER 62
+ R
Sbjct: 975 SCSR 964
>BQ145145 similar to SP|O64407|AMYB_ Beta-amylase (EC 3.2.1.2) (1
4-alpha-D-glucan maltohydrolase). [Cowpea] {Vigna
unguiculata}, partial (7%)
Length = 283
Score = 24.3 bits (51), Expect = 8.2
Identities = 21/63 (33%), Positives = 29/63 (45%), Gaps = 8/63 (12%)
Frame = -1
Query: 44 IPRMNLIPSGANVSITFERRQFPLVLSFAMTINKSQG--------QTLTSVGLYLPRPVF 95
I +N+I SG N SIT + S M++++SQG TS G +PR F
Sbjct: 190 IREVNIISSGFNNSITILPDELGSQPSTFMSVSESQGNGGIGLVASLRTSTG--IPRAEF 17
Query: 96 THG 98
G
Sbjct: 16 GRG 8
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.141 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,907,842
Number of Sequences: 36976
Number of extensions: 33324
Number of successful extensions: 172
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 171
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 172
length of query: 102
length of database: 9,014,727
effective HSP length: 78
effective length of query: 24
effective length of database: 6,130,599
effective search space: 147134376
effective search space used: 147134376
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 50 (23.9 bits)
Medicago: description of AC148485.5


