

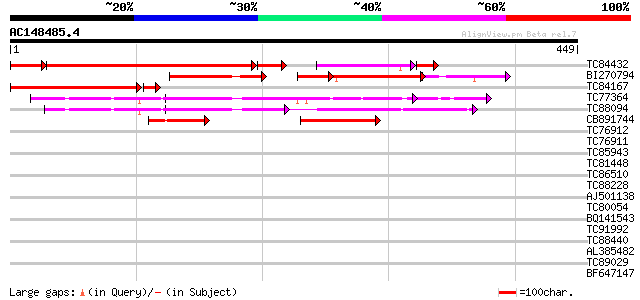
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148485.4 - phase: 0
(449 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84432 similar to GP|9757999|dbj|BAB08421.1 polypyrimidine trac... 257 9e-84
BI270794 similar to GP|18921326|gb putative polypyrimidine tract... 130 3e-49
TC84167 homologue to GP|9757999|dbj|BAB08421.1 polypyrimidine tr... 160 6e-41
TC77364 similar to GP|20160852|dbj|BAB89791. contains EST AU0557... 144 5e-35
TC88094 similar to GP|20160852|dbj|BAB89791. contains EST AU0557... 128 5e-30
CB891744 similar to GP|18921326|gb| putative polypyrimidine trac... 97 1e-20
TC76912 homologue to GP|13543234|gb|AAH05782.1 Unknown (protein ... 40 0.002
TC76911 similar to GP|13543234|gb|AAH05782.1 Unknown (protein fo... 40 0.002
TC85943 similar to PIR|T07080|T07080 shock protein SRC2 - soybea... 39 0.003
TC81448 weakly similar to OMNI|NT01MC3624 toxin secretion ATP-bi... 39 0.005
TC86510 similar to GP|20160777|dbj|BAB89718. putative ribonucleo... 38 0.009
TC88228 similar to GP|13605916|gb|AAK32943.1 AT4g00830/A_TM018A1... 37 0.012
AJ501138 similar to GP|14021481|dbj unknown protein {Mesorhizobi... 37 0.012
TC80054 SP|Q28009|FUS_BOVIN RNA-binding protein FUS (Pigpen prot... 36 0.035
BQ141543 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 35 0.045
TC91992 similar to GP|8099125|dbj|BAA90497.1 rice EST C27893 cor... 35 0.077
TC88440 weakly similar to GP|7293965|gb|AAF49324.1| Eip74EF gene... 34 0.13
AL385482 similar to GP|5106924|gb|A putative cell wall protein {... 33 0.17
TC89029 weakly similar to GP|15450467|gb|AAK96527.1 At1g12810/F1... 33 0.17
BF647147 weakly similar to GP|7303636|gb|A CG13214-PA {Drosophil... 33 0.17
>TC84432 similar to GP|9757999|dbj|BAB08421.1 polypyrimidine tract-binding
RNA transport protein-like {Arabidopsis thaliana},
partial (50%)
Length = 685
Score = 257 bits (656), Expect(3) = 9e-84
Identities = 123/166 (74%), Positives = 149/166 (89%)
Frame = +3
Query: 30 SDEELADLCSPFGKVMSTKCNVGVNQNQAFVQFAELNQAISMVSYYASSSEPAQVRGKTV 89
S+EEL +L +PFGKV++TKCNVG N+NQAF++FA+LNQAI+M+SYYASSSEPAQVRGKTV
Sbjct: 114 SEEELIELGNPFGKVVNTKCNVGSNRNQAFIEFADLNQAIAMISYYASSSEPAQVRGKTV 293
Query: 90 YIQYSNRHHIVNSNSPGDIPGNVLLVTIEGVEAGDVSIDVIHLVFSAFGFVHKLATFEKT 149
Y+QYSNR IV++ + D+ GNVLLVT+EG +A VSIDV+HLVFSAFGFVHK+ TFEKT
Sbjct: 294 YLQYSNRQEIVHNKTAADVAGNVLLVTVEGEDARLVSIDVLHLVFSAFGFVHKITTFEKT 473
Query: 150 AGFQALIQYTDAETAASAKDSLDGRSIPRYLLPEHVGACNLRISYS 195
AGFQAL+Q++DAETA SAKD+LDGRSIPRYLL EHVG C L+I+YS
Sbjct: 474 AGFQALVQFSDAETATSAKDALDGRSIPRYLLSEHVGPCTLKITYS 611
Score = 64.7 bits (156), Expect(2) = 1e-12
Identities = 38/85 (44%), Positives = 48/85 (55%), Gaps = 7/85 (8%)
Frame = +3
Query: 244 NVLLATIENMQYA-VPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKET 302
NVLL T+E V +DVLH VFSAFGFV K+ F+K AL+Q+ D A +AK+
Sbjct: 357 NVLLVTVEGEDARLVSIDVLHLVFSAFGFVHKITTFEKTAGFQALVQFSDAETATSAKDA 536
Query: 303 LEGHCI------YDGGYCKLHLTYS 321
L+G I G C L +TYS
Sbjct: 537 LDGRSIPRYLLSEHVGPCTLKITYS 611
Score = 50.1 bits (118), Expect(3) = 9e-84
Identities = 22/29 (75%), Positives = 23/29 (78%)
Frame = +2
Query: 1 MSSSKQQQQFRCTQIPSKVLHFRNLPWEC 29
M+S QQQFR TQ PSKVLH RNLPWEC
Sbjct: 26 MASVSSQQQFRYTQPPSKVLHLRNLPWEC 112
Score = 43.1 bits (100), Expect(3) = 9e-84
Identities = 18/23 (78%), Positives = 20/23 (86%)
Frame = +1
Query: 197 HRDLNIKFQSNRSRDYTNPMLPV 219
H DL +KFQS+RSRDYTNP LPV
Sbjct: 616 HSDLTVKFQSHRSRDYTNPYLPV 684
Score = 25.8 bits (55), Expect(2) = 1e-12
Identities = 11/17 (64%), Positives = 13/17 (75%)
Frame = +1
Query: 323 HTDLNVKAFSDKSRDYT 339
H+DL VK S +SRDYT
Sbjct: 616 HSDLTVKFQSHRSRDYT 666
>BI270794 similar to GP|18921326|gb putative polypyrimidine tract-binding
protein {Oryza sativa}, partial (26%)
Length = 659
Score = 130 bits (327), Expect(3) = 3e-49
Identities = 61/79 (77%), Positives = 71/79 (89%), Gaps = 3/79 (3%)
Frame = +3
Query: 254 QYAV---PLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKETLEGHCIYD 310
+YAV P+DV+++VFSAFG VQK+AMF+KNG T ALIQYPD+T AAAA+E LEGHCIYD
Sbjct: 72 EYAVCLLPVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREALEGHCIYD 251
Query: 311 GGYCKLHLTYSRHTDLNVK 329
GGYCKLHL+YSRHTDLNVK
Sbjct: 252 GGYCKLHLSYSRHTDLNVK 308
Score = 75.1 bits (183), Expect = 5e-14
Identities = 40/77 (51%), Positives = 50/77 (63%)
Frame = +3
Query: 127 IDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPEHVG 186
+DVI+ VFSAFG V K+A FEK QALIQY D TAA+A+++L+G I G
Sbjct: 96 VDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREALEGHCI------YDGG 257
Query: 187 ACNLRISYSAHRDLNIK 203
C L +SYS H DLN+K
Sbjct: 258 YCKLHLSYSRHTDLNVK 308
Score = 53.5 bits (127), Expect(3) = 3e-49
Identities = 33/89 (37%), Positives = 38/89 (42%), Gaps = 20/89 (22%)
Frame = +2
Query: 328 VKAFSDKSRDYTVLDPSLHAAQAPAWQTTQAATMYSGSM--------------------G 367
+ AFSDKSRDYTV AP WQ QAA MY + G
Sbjct: 404 LNAFSDKSRDYTV-----PLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGG 568
Query: 368 QMPSWDPNQQEVTQSYLSAPGTFPSGQAA 396
Q+PSWD Q V Y+ PG +P A
Sbjct: 569 QVPSWDLTQHAVRPGYVPVPGAYPGQTGA 655
Score = 50.4 bits (119), Expect(3) = 3e-49
Identities = 24/28 (85%), Positives = 26/28 (92%)
Frame = +1
Query: 229 QPAIGPDGKRKEHKSNVLLATIENMQYA 256
QPAIGPDGKR E +SNVLLA+IENMQYA
Sbjct: 4 QPAIGPDGKRIETESNVLLASIENMQYA 87
>TC84167 homologue to GP|9757999|dbj|BAB08421.1 polypyrimidine tract-binding
RNA transport protein-like {Arabidopsis thaliana},
partial (27%)
Length = 543
Score = 160 bits (406), Expect(2) = 6e-41
Identities = 75/104 (72%), Positives = 89/104 (85%)
Frame = +1
Query: 1 MSSSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFV 60
MSS Q QFR TQ PSKVLH RNLPWEC +EEL +L PFGKV++TKCNVG N+NQAF+
Sbjct: 184 MSSVSTQPQFRYTQPPSKVLHLRNLPWECIEEELIELGKPFGKVVNTKCNVGANRNQAFI 363
Query: 61 QFAELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNS 104
+FA++NQAI+M+SYYASSSEPAQVRGKTVY+QYSNR IVN+ +
Sbjct: 364 EFADINQAIAMISYYASSSEPAQVRGKTVYLQYSNRQEIVNNQN 495
Score = 25.0 bits (53), Expect(2) = 6e-41
Identities = 10/13 (76%), Positives = 12/13 (91%)
Frame = +2
Query: 107 DIPGNVLLVTIEG 119
D+ GNVL+VTIEG
Sbjct: 503 DVAGNVLVVTIEG 541
>TC77364 similar to GP|20160852|dbj|BAB89791. contains EST
AU055715(S20012)~similar to nuclear
ribonucleoprotein~unknown protein, partial (50%)
Length = 2480
Score = 144 bits (364), Expect = 5e-35
Identities = 107/343 (31%), Positives = 171/343 (49%), Gaps = 36/343 (10%)
Frame = +2
Query: 17 SKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQFAELNQAISMVSYYA 76
SKV+H RN+ E S+ +L L PFG V TK + +NQA VQ ++ A S + +Y
Sbjct: 743 SKVIHVRNVGHEISENDLLQLFQPFGVV--TKLVMLRAKNQALVQMQDVASADSAIQFYI 916
Query: 77 SSSEPAQVRGKTVYIQYSNRHHIVN---SNSPGDIPGNVLLVTIEGVEAGDVSIDVIHLV 133
+S +RG+ VY+Q+S+ + + D P +LLVT+ V +++DV+ V
Sbjct: 917 NSQP--SIRGRNVYVQFSSHRELTTMDQNQGREDEPNRILLVTVHQV-LYPMTVDVLQQV 1087
Query: 134 FSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPEHVGACNLRIS 193
FS GFV K+ TF+K+AGFQALIQY ++A +A+ +L GR++ + G C L I
Sbjct: 1088 FSPHGFVEKIVTFQKSAGFQALIQYETRQSAVTARGALQGRNV-------YDGCCQLDIQ 1246
Query: 194 YSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDS--------------------------- 226
+S +L + + ++RSRD+TNP LP +Q S
Sbjct: 1247 FSNLDELQVNYNNDRSRDFTNPNLPTDQKGRPSQAGYGDAGNYGGIQGSGPRQAGFSQMG 1426
Query: 227 ---ALQPAIG---PDGKRKEHKSNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDK 280
A++ A G P G ++ +L T N + D L ++FS +G + ++ + +
Sbjct: 1427 NAQAIEAAFGGDLPPGITGTNERCTILVTNLNPD-RIDEDKLFNLFSIYGNIVRIKLL-R 1600
Query: 281 NGHTHALIQYPDLTIAAAAKETLEGHCIYDGGYCKLHLTYSRH 323
N HAL+Q D A A L+G ++ L + +S+H
Sbjct: 1601 NKPDHALVQMGDGFQAELAVHFLKGAMLFGK---HLDVNFSKH 1720
Score = 86.7 bits (213), Expect = 2e-17
Identities = 75/259 (28%), Positives = 119/259 (44%), Gaps = 1/259 (0%)
Frame = +2
Query: 124 DVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPE 183
++S + + +F FG V KL A QAL+Q D +A SA I Y+ +
Sbjct: 776 EISENDLLQLFQPFGVVTKLVMLR--AKNQALVQMQDVASADSA--------IQFYINSQ 925
Query: 184 -HVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDGKRKEHK 242
+ N+ + +S+HR+L Q NQ R++
Sbjct: 926 PSIRGRNVYVQFSSHRELTTMDQ--------------NQG---------------REDEP 1018
Query: 243 SNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKET 302
+ +LL T+ + Y + +DVL VFS GFV+K+ F K+ ALIQY A A+
Sbjct: 1019NRILLVTVHQVLYPMTVDVLQQVFSPHGFVEKIVTFQKSAGFQALIQYETRQSAVTARGA 1198
Query: 303 LEGHCIYDGGYCKLHLTYSRHTDLNVKAFSDKSRDYTVLDPSLHAAQAPAWQTTQAATMY 362
L+G +YD G C+L + +S +L V +D+SRD+T +P+L Q + +QA
Sbjct: 1199LQGRNVYD-GCCQLDIQFSNLDELQVNYNNDRSRDFT--NPNLPTDQ--KGRPSQAGYGD 1363
Query: 363 SGSMGQMPSWDPNQQEVTQ 381
+G+ G + P Q +Q
Sbjct: 1364AGNYGGIQGSGPRQAGFSQ 1420
>TC88094 similar to GP|20160852|dbj|BAB89791. contains EST
AU055715(S20012)~similar to nuclear
ribonucleoprotein~unknown protein, partial (22%)
Length = 890
Score = 128 bits (321), Expect = 5e-30
Identities = 73/197 (37%), Positives = 116/197 (58%), Gaps = 3/197 (1%)
Frame = +1
Query: 28 ECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQFAELNQAISMVSYYASSSEPAQVRGK 87
E S+ +L L PFG + TK + +NQA +Q ++ A+S + +Y ++ +RG+
Sbjct: 130 EISENDLLQLFQPFGVI--TKLVMLRAKNQALIQMQDVPSAVSALHFYGNTQP--SIRGR 297
Query: 88 TVYIQYSNRHHIVN---SNSPGDIPGNVLLVTIEGVEAGDVSIDVIHLVFSAFGFVHKLA 144
VY+Q+S+ + S GD P +LLVTI V +++DV++ VFS G V K+
Sbjct: 298 NVYVQFSSHQELTTIDQSQGRGDEPNRILLVTIHHV-LYPITVDVLYQVFSPHGSVEKIV 474
Query: 145 TFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPEHVGACNLRISYSAHRDLNIKF 204
TF+K+AGFQALIQY +++ +A+ +L GR+I + G C L I +S +L + +
Sbjct: 475 TFQKSAGFQALIQYQSQQSSITARTALQGRNI-------YDGCCQLDIQFSNLDELQVNY 633
Query: 205 QSNRSRDYTNPMLPVNQ 221
++RSRD+TNP LP Q
Sbjct: 634 NNDRSRDFTNPNLPTEQ 684
Score = 82.0 bits (201), Expect = 4e-16
Identities = 72/248 (29%), Positives = 109/248 (43%), Gaps = 1/248 (0%)
Frame = +1
Query: 124 DVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPE 183
++S + + +F FG + KL A QALIQ D +A SA G + P
Sbjct: 130 EISENDLLQLFQPFGVITKLVMLR--AKNQALIQMQDVPSAVSALHFY-GNTQPS----- 285
Query: 184 HVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDGKRKEHKS 243
+ N+ + +S+H++L QS D N
Sbjct: 286 -IRGRNVYVQFSSHQELTTIDQSQGRGDEPN----------------------------- 375
Query: 244 NVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKETL 303
+LL TI ++ Y + +DVL+ VFS G V+K+ F K+ ALIQY + A+ L
Sbjct: 376 RILLVTIHHVLYPITVDVLYQVFSPHGSVEKIVTFQKSAGFQALIQYQSQQSSITARTAL 555
Query: 304 EGHCIYDGGYCKLHLTYSRHTDLNVKAFSDKSRDYTVLD-PSLHAAQAPAWQTTQAATMY 362
+G IYD G C+L + +S +L V +D+SRD+T + P+ + P A MY
Sbjct: 556 QGRNIYD-GCCQLDIQFSNLDELQVNYNNDRSRDFTNPNLPTEQKGRPPQLGYGDAGNMY 732
Query: 363 SGSMGQMP 370
G G P
Sbjct: 733 -GVQGSGP 753
>CB891744 similar to GP|18921326|gb| putative polypyrimidine tract-binding
protein {Oryza sativa}, partial (13%)
Length = 366
Score = 97.4 bits (241), Expect = 1e-20
Identities = 47/63 (74%), Positives = 56/63 (88%)
Frame = +1
Query: 231 AIGPDGKRKEHKSNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQY 290
AIGPDGKR E +SNVLLA+IENMQYAV +DV+++VFSAFG VQK+AMF+KNG T ALIQY
Sbjct: 1 AIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQY 180
Query: 291 PDL 293
P +
Sbjct: 181 PGI 189
Score = 51.6 bits (122), Expect = 6e-07
Identities = 28/48 (58%), Positives = 35/48 (72%)
Frame = +1
Query: 111 NVLLVTIEGVEAGDVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQY 158
NVLL +IE ++ V++DVI+ VFSAFG V K+A FEK QALIQY
Sbjct: 40 NVLLASIENMQYA-VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQY 180
>TC76912 homologue to GP|13543234|gb|AAH05782.1 Unknown (protein for
MGC:12025) {Mus musculus}, partial (26%)
Length = 1077
Score = 39.7 bits (91), Expect = 0.002
Identities = 19/49 (38%), Positives = 23/49 (46%), Gaps = 6/49 (12%)
Frame = +2
Query: 387 PGTFPSGQAAPPFPGYSPA------AVPPAGASPHSHMPPSSFAGAFPG 429
PG +P Q PP GY PA PP+G P PP+ + GA G
Sbjct: 167 PGAYPPQQGYPPQQGYPPAGYPPQQGYPPSGYPPQQGYPPAGYPGAHAG 313
>TC76911 similar to GP|13543234|gb|AAH05782.1 Unknown (protein for
MGC:12025) {Mus musculus}, partial (57%)
Length = 1065
Score = 39.7 bits (91), Expect = 0.002
Identities = 19/49 (38%), Positives = 23/49 (46%), Gaps = 6/49 (12%)
Frame = +2
Query: 387 PGTFPSGQAAPPFPGYSPA------AVPPAGASPHSHMPPSSFAGAFPG 429
PG +P Q PP GY PA PP+G P PP+ + GA G
Sbjct: 233 PGAYPPQQGYPPQQGYPPAGYPPQQGYPPSGYPPQQGYPPAGYPGAHAG 379
>TC85943 similar to PIR|T07080|T07080 shock protein SRC2 - soybean, partial
(74%)
Length = 1279
Score = 39.3 bits (90), Expect = 0.003
Identities = 36/136 (26%), Positives = 54/136 (39%), Gaps = 11/136 (8%)
Frame = +3
Query: 322 RHTDLNVKAFSDKSRDYTVLDPSL-HAAQAPAWQTTQAATMYSGSMGQMPSWDPNQQEVT 380
R + V+ S K++ L QAPA +TT G N+ E
Sbjct: 459 RQVNYQVRTSSGKAKGNLNLSYKFGEKVQAPAMKTTSKHEPVKGK---------NEHEPV 611
Query: 381 QSYLSAPGTFPSGQAAPPF----------PGYSPAAVPPAGASPHSHMPPSSFAGAFPGS 430
+Y PG +G ++ P+ PGY PPA A+ + PP+ G +P +
Sbjct: 612 MAY-PPPGMAAAGSSSVPYGTPYPPQPQQPGYGYPPAPPAAAA-YGGYPPAQ-PGGYPPA 782
Query: 431 QPHYGWESRFRLNYNP 446
QP YG+ + Y P
Sbjct: 783 QPGYGYPPQQGHGYPP 830
>TC81448 weakly similar to OMNI|NT01MC3624 toxin secretion ATP-binding
protein putative {Magnetococcus sp. MC-1}, partial (4%)
Length = 655
Score = 38.5 bits (88), Expect = 0.005
Identities = 25/49 (51%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Frame = -2
Query: 388 GTFPSGQAAPPFPGYSPAAVPP-AGASPHSHMPPSSFAGAFPGSQPHYG 435
G FPS PP G PA PP AGA P PP + AGAFP S P G
Sbjct: 240 GAFPS----PPGAGALPAGAPPGAGAFPS---PPGAGAGAFPASPPGAG 115
Score = 28.5 bits (62), Expect = 5.5
Identities = 20/42 (47%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Frame = -2
Query: 388 GTFPSGQAAPPFPGYS-PAAVPPAGASPHSHMPPSSFAGAFP 428
G P G A PG + PA+ P AGA P + PP AGAFP
Sbjct: 345 GVTPGGPPAGGVPGAAFPASPPGAGALP-AGAPPG--AGAFP 229
Score = 27.7 bits (60), Expect = 9.4
Identities = 17/48 (35%), Positives = 21/48 (43%), Gaps = 7/48 (14%)
Frame = +2
Query: 386 APGTFPSGQAAPP-------FPGYSPAAVPPAGASPHSHMPPSSFAGA 426
APG P+G+A P PG PA PP S PP++ A
Sbjct: 242 APGGAPAGKAPAPGGDAGKAAPGTPPAGGPPGVTPSGSASPPAATPAA 385
>TC86510 similar to GP|20160777|dbj|BAB89718. putative ribonucleoprotein
{Oryza sativa (japonica cultivar-group)}, partial (67%)
Length = 1587
Score = 37.7 bits (86), Expect = 0.009
Identities = 32/108 (29%), Positives = 47/108 (42%), Gaps = 4/108 (3%)
Frame = +3
Query: 3 SSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNV----GVNQNQA 58
SSK + P L ++P E D+ELA+ PFG+V+S K V GV++
Sbjct: 1149 SSKNSGGGQAEGPPGANLFIYHIPQEFGDQELANAFQPFGRVLSAKVFVDKATGVSKCFG 1328
Query: 59 FVQFAELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNSPG 106
FV + A S + S Q+ GK + +Q+ N PG
Sbjct: 1329 FVSYDSPEAAQSAI----SMMNGCQLGGKKLKVQHK------RDNKPG 1442
Score = 27.7 bits (60), Expect = 9.4
Identities = 16/59 (27%), Positives = 21/59 (35%)
Frame = +3
Query: 388 GTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFPGSQPHYGWESRFRLNYNP 446
G P G PP+ GY A G P+ PP + PH + R + P
Sbjct: 792 GAMPMGYV-PPYNGYGYQAPGSYGLMPYRMPPPMQNQPGYHNMMPHMNQGNALRPDLGP 965
>TC88228 similar to GP|13605916|gb|AAK32943.1 AT4g00830/A_TM018A10_14
{Arabidopsis thaliana}, partial (67%)
Length = 1712
Score = 37.4 bits (85), Expect = 0.012
Identities = 29/111 (26%), Positives = 48/111 (43%), Gaps = 5/111 (4%)
Frame = +1
Query: 25 LPWECSDEELADLCSPFGKVMSTKC----NVGVNQNQAFVQFAELNQAISMVSYYASSSE 80
LP + SD+++ +LC P G ++ K G ++ AFV + A +
Sbjct: 484 LPRDTSDDDVRELCEPMGDIVEIKLIKDRETGESKGYAFVGYKTKEVAQKAI----DDIH 651
Query: 81 PAQVRGKTVYIQYS-NRHHIVNSNSPGDIPGNVLLVTIEGVEAGDVSIDVI 130
+ +GKT+ S +H + N P + +EGV G SID+I
Sbjct: 652 NKEFKGKTLRCLLSETKHRLFIGNIPKTWTEDEFRKAVEGVGPGVESIDLI 804
>AJ501138 similar to GP|14021481|dbj unknown protein {Mesorhizobium loti},
partial (16%)
Length = 544
Score = 37.4 bits (85), Expect = 0.012
Identities = 28/89 (31%), Positives = 43/89 (47%), Gaps = 1/89 (1%)
Frame = -3
Query: 343 PSLHAAQAPAWQTTQAATMYSGSMGQMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFPGY 402
P+ A AP T ++++ ++G +PS + L + G PS A P +
Sbjct: 533 PTRPATTAPV--TKPLSSLFPAAVGALPSAGAFP---SAGALPSAGALPSAGALPSAGAF 369
Query: 403 -SPAAVPPAGASPHSHMPPSSFAGAFPGS 430
S A+P AGA P + PS+ AGAFP +
Sbjct: 368 PSAGALPSAGALPSAGAFPSAGAGAFPSA 282
>TC80054 SP|Q28009|FUS_BOVIN RNA-binding protein FUS (Pigpen protein).
[Bovine] {Bos taurus}, partial (4%)
Length = 988
Score = 35.8 bits (81), Expect = 0.035
Identities = 31/99 (31%), Positives = 38/99 (38%), Gaps = 6/99 (6%)
Frame = +1
Query: 343 PSLHAAQAPAWQTTQAATMYSGSMGQMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPF--P 400
P AAQ + T Y + P Q + A G S AA + P
Sbjct: 388 PPQGAAQPSYGVPPTSQTAYGSQPQTQSGYGPPQTQKPSGTPPAYGQSQSPNAAGGYGQP 567
Query: 401 GYSPAAVPPAGAS----PHSHMPPSSFAGAFPGSQPHYG 435
GY P+ PP+G S P S PPS + SQP YG
Sbjct: 568 GYPPSQPPPSGYSQPGYPPSQPPPSGY------SQPGYG 666
>BQ141543 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (15%)
Length = 1271
Score = 35.4 bits (80), Expect = 0.045
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 1/99 (1%)
Frame = -2
Query: 317 HLTYSRHTDLNVKAFSDKSRDYTVLDPSLHAAQAPAWQTTQAATMYSGSMGQMPSWDPNQ 376
H+ H+ ++ + S SR L P H A AP + +YS S+ S+
Sbjct: 907 HIATRCHSSVSTRPAS--SRATRPLLPLSHRAPAPPYHGVIHFALYSHSISGYISYGSYW 734
Query: 377 QEVTQSYLSAPGTFP-SGQAAPPFPGYSPAAVPPAGASP 414
S+ A T P SG +PP P + P +PP+ A P
Sbjct: 733 VFSPVSFRLAS*TIPPSGSPSPPPPSHPPRHLPPSPALP 617
>TC91992 similar to GP|8099125|dbj|BAA90497.1 rice EST C27893 corresponds to
a region of the predicated gene; unknown protein {Oryza
sativa}, partial (28%)
Length = 733
Score = 34.7 bits (78), Expect = 0.077
Identities = 20/54 (37%), Positives = 25/54 (46%), Gaps = 4/54 (7%)
Frame = +1
Query: 387 PGTFPSGQAAPPFPGYSP---AAVPPAGASPHSHMPPSSFAGAFPGS-QPHYGW 436
P P+ + PP GY P A PP G S PP + G +P S PH G+
Sbjct: 121 PAAPPASYSYPPQGGYYPPPQTAYPPQGGYYPSQYPPHAAGGGYPPSGYPHSGY 282
>TC88440 weakly similar to GP|7293965|gb|AAF49324.1| Eip74EF gene product
{Drosophila melanogaster}, partial (5%)
Length = 711
Score = 33.9 bits (76), Expect = 0.13
Identities = 18/40 (45%), Positives = 21/40 (52%)
Frame = +1
Query: 387 PGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGA 426
P P A PP PA PPAGA+P + P SS +GA
Sbjct: 256 PAGGPPAGATPPAGATPPAGGPPAGATPPAGGPSSSPSGA 375
Score = 30.8 bits (68), Expect = 1.1
Identities = 17/34 (50%), Positives = 19/34 (55%)
Frame = +1
Query: 395 AAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFP 428
A PP G A PPAGA+P + PP AGA P
Sbjct: 247 ATPPAGGPPAGATPPAGATPPAGGPP---AGATP 339
Score = 28.9 bits (63), Expect = 4.2
Identities = 16/36 (44%), Positives = 17/36 (46%)
Frame = +1
Query: 393 GQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFP 428
G A P +P A PPAG P PP AGA P
Sbjct: 208 GDATPVGGDATPGATPPAGGPPAGATPP---AGATP 306
>AL385482 similar to GP|5106924|gb|A putative cell wall protein {Medicago
truncatula}, partial (42%)
Length = 402
Score = 33.5 bits (75), Expect = 0.17
Identities = 19/43 (44%), Positives = 21/43 (48%)
Frame = +2
Query: 386 APGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFP 428
AP P G+ APP G +P PP GA P PP A A P
Sbjct: 29 APKDAPKGKVAPPPAGGAP---PPGGAPPAGGAPPPGGAPAGP 148
>TC89029 weakly similar to GP|15450467|gb|AAK96527.1 At1g12810/F13K23_4
{Arabidopsis thaliana}, partial (71%)
Length = 643
Score = 33.5 bits (75), Expect = 0.17
Identities = 15/38 (39%), Positives = 17/38 (44%), Gaps = 3/38 (7%)
Frame = +3
Query: 398 PFPGYSPAAVPPAGASPHSH---MPPSSFAGAFPGSQP 432
P PG+ PP G PH H PP + G FP P
Sbjct: 66 PPPGFGSPYPPPPGPPPHQHHEGYPPPGYPGGFPPPPP 179
>BF647147 weakly similar to GP|7303636|gb|A CG13214-PA {Drosophila
melanogaster}, partial (8%)
Length = 426
Score = 33.5 bits (75), Expect = 0.17
Identities = 15/38 (39%), Positives = 17/38 (44%), Gaps = 3/38 (7%)
Frame = +3
Query: 398 PFPGYSPAAVPPAGASPHSH---MPPSSFAGAFPGSQP 432
P PG+ PP G PH H PP + G FP P
Sbjct: 39 PPPGFGSPYPPPPGPPPHQHHEGYPPPGYPGGFPPPPP 152
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.131 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,868,075
Number of Sequences: 36976
Number of extensions: 201695
Number of successful extensions: 1808
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 1514
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1715
length of query: 449
length of database: 9,014,727
effective HSP length: 99
effective length of query: 350
effective length of database: 5,354,103
effective search space: 1873936050
effective search space used: 1873936050
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148485.4


