

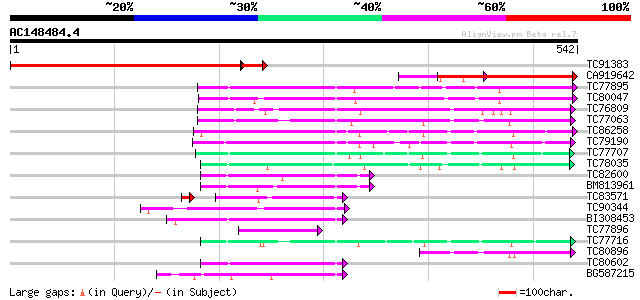
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.4 - phase: 0
(542 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91383 similar to GP|9858782|gb|AAG01129.1| BAC19.14 {Lycopersi... 467 e-141
CA919642 similar to PIR|E85433|E854 SCARECROW-like protein [impo... 269 2e-72
TC77895 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 ... 202 3e-52
TC80047 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 ... 180 1e-45
TC76809 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 ... 105 6e-23
TC77063 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 ... 100 2e-21
TC86258 similar to GP|16930433|gb|AAL31902.1 AT4g17230/dl4650c {... 90 3e-18
TC79190 similar to PIR|E96542|E96542 scarecrow-like protein [imp... 81 1e-15
TC77707 weakly similar to GP|8777405|dbj|BAA96995.1 SCARECROW ge... 73 3e-13
TC78035 similar to GP|8778540|gb|AAF79548.1| F22G5.9 {Arabidopsi... 70 2e-12
TC82600 similar to GP|9757937|dbj|BAB08425.1 SCARECROW gene regu... 69 5e-12
BM813961 homologue to GP|13365610|db SCARECROW {Pisum sativum}, ... 68 8e-12
TC83571 similar to PIR|T10552|T10552 hypothetical protein T12G13... 65 2e-11
TC90344 weakly similar to PIR|H86282|H86282 protein F10B6.34 [im... 66 4e-11
BI308453 weakly similar to PIR|T02736|T0 probable SCARECROW gene... 60 2e-09
TC77896 weakly similar to GP|22087131|gb|AAM90848.1 hairy merist... 57 2e-08
TC77716 similar to GP|20465587|gb|AAM20276.1 unknown protein {Ar... 57 2e-08
TC80896 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 ... 56 4e-08
TC80602 similar to GP|14517552|gb|AAK62666.1 F17J6.12/F17J6.12 {... 55 9e-08
BG587215 similar to GP|8453100|gb| short-root protein {Arabidops... 50 3e-06
>TC91383 similar to GP|9858782|gb|AAG01129.1| BAC19.14 {Lycopersicon
esculentum}, partial (11%)
Length = 819
Score = 467 bits (1202), Expect(2) = e-141
Identities = 223/225 (99%), Positives = 224/225 (99%)
Frame = +2
Query: 1 MRVPPPAVSSSSSSPQPKLTTTQNIVVNVPTPNPTPTPTTTTLCYEPRSVLELCRSPSPE 60
MRVPPPAVSSSSSSPQPKLTTTQNIVVNVPTPNPTPTPTTTTLCYEPRSVLELCRSPSPE
Sbjct: 80 MRVPPPAVSSSSSSPQPKLTTTQNIVVNVPTPNPTPTPTTTTLCYEPRSVLELCRSPSPE 259
Query: 61 KKPSQQEQEHNPEVEQEDHASLPNLDWWDSIMKDLGLQDDSSTPIIPLLKNTNNTSEIYP 120
KKPSQQEQEHNPEVEQEDHASLPNLDWWDSIMKDLGLQDDSSTPIIPLLKNTNNTSEIYP
Sbjct: 260 KKPSQQEQEHNPEVEQEDHASLPNLDWWDSIMKDLGLQDDSSTPIIPLLKNTNNTSEIYP 439
Query: 121 NPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFI 180
NPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFI
Sbjct: 440 NPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFI 619
Query: 181 EELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFK 225
EELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAF F+
Sbjct: 620 EELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFSFQ 754
Score = 53.9 bits (128), Expect(2) = e-141
Identities = 25/25 (100%), Positives = 25/25 (100%)
Frame = +3
Query: 222 FHFKDALQSLLSGSNRTNPPRLSSM 246
FHFKDALQSLLSGSNRTNPPRLSSM
Sbjct: 744 FHFKDALQSLLSGSNRTNPPRLSSM 818
>CA919642 similar to PIR|E85433|E854 SCARECROW-like protein [imported] -
Arabidopsis thaliana, partial (12%)
Length = 661
Score = 269 bits (687), Expect = 2e-72
Identities = 133/133 (100%), Positives = 133/133 (100%)
Frame = -2
Query: 410 FVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKII 469
FVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKII
Sbjct: 546 FVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKII 367
Query: 470 AAVEAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFW 529
AAVEAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFW
Sbjct: 366 AAVEAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFW 187
Query: 530 HERAMVATSAWRC 542
HERAMVATSAWRC
Sbjct: 186 HERAMVATSAWRC 148
Score = 75.9 bits (185), Expect = 4e-14
Identities = 49/93 (52%), Positives = 55/93 (58%), Gaps = 7/93 (7%)
Frame = -1
Query: 372 DGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVF-----VDGEGWTEAAAAASFR 426
DGEKTAIL PAIFCRLGSEGTAAFLSDVRRITPGVVVF +DG
Sbjct: 661 DGEKTAIL*RPAIFCRLGSEGTAAFLSDVRRITPGVVVFR*R*RMDG----------GCS 512
Query: 427 RGVVNS--LEFYSMMLESLDASVAAGGGGEWAR 457
GV +S E + ++L+ GGGG R
Sbjct: 511 SGVFSSWCCE*FGVLLDDAGVIGCVGGGGRRGR 413
>TC77895 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 {Arabidopsis
thaliana}, partial (41%)
Length = 2591
Score = 202 bits (514), Expect = 3e-52
Identities = 131/368 (35%), Positives = 206/368 (55%), Gaps = 5/368 (1%)
Frame = +3
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
I++L +AA+ D + + AQ IL RLN ++ SP GKP RAAF+FK+ALQ LL + +
Sbjct: 1314 IDQLFKAAELIDAGNPEHAQGILARLNHQI-SPMGKPFQRAAFYFKEALQLLLQSNVNNS 1490
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQ 299
S ++ I +K+FS ISP+ F+ FT+NQALLEA+ G +H++DF+IG G Q
Sbjct: 1491 NNNSFSPTSLLLKIGAYKSFSEISPVLQFANFTSNQALLEAVEGFDRIHIIDFDIGFGGQ 1670
Query: 300 YASLMKEIAEKAVNGSPLLRITAVVPEEY--AVESRLIRENLNQFAHDLGIRVQVDFVPL 357
++S M+E+A + G+P L+ITA V + +E ENL Q+A ++ + +++ + L
Sbjct: 1671 WSSFMQELALRN-GGAPALKITAFVSPSHHDEIELSFTNENLMQYAGEINMSFELEILTL 1847
Query: 358 RTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWT 417
+ +VS+ D E + L F S L V+++ P +VV +D +
Sbjct: 1848 ESLNSVSWPQ-PLRDCEAVVVNLPICSFSNYPSY-LPLVLRFVKQLMPKIVVTLDR---S 2012
Query: 418 EAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRP---KIIAAVEA 474
A F + +V +L+ YS +LESLDA V+ + + IE L+P K++
Sbjct: 2013 CDRTDAPFPQHMVFALQSYSGLLESLDA-VSVNVHPDVLQMIEKYYLQPAIEKLVLGRLR 2189
Query: 475 AGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAM 534
+ RT PW+ +G P+ S F + QAECL+ ++ RGFHV K+Q LVL W + +
Sbjct: 2190 SQDRTLPWKNLLLSSGFSPLTFSNFTESQAECLVQRIPGRGFHVEKKQNSLVLCWQRKDL 2369
Query: 535 VATSAWRC 542
++ S WRC
Sbjct: 2370 ISVSTWRC 2393
>TC80047 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20
{Arabidopsis thaliana}, partial (30%)
Length = 1357
Score = 180 bits (456), Expect = 1e-45
Identities = 122/370 (32%), Positives = 193/370 (51%), Gaps = 8/370 (2%)
Frame = +2
Query: 181 EELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLL--SGSNRT 238
++L + A+ + + AQ IL RLN +L SP G P RA+F+ K+ALQ +L +G+N T
Sbjct: 44 DQLFKTAELIEAGNPVQAQGILARLNHQL-SPIGNPFQRASFYMKEALQLMLHSNGNNLT 220
Query: 239 NPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGI 298
S + + I +K+FS ISP+ F+ FT NQ+L+EAL +HV+DF+IG G+
Sbjct: 221 ----AFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGV 388
Query: 299 QYASLMKEIAEKAVNGSPLLRITAVVPEEYA--VESRLIRENLNQFAHDLGIRVQVDFVP 356
Q++S M+EI ++ NG P L+ITAVV +E +ENL+Q+A DL I + + +
Sbjct: 389 QWSSFMQEIVLRS-NGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLN 565
Query: 357 LRTFETVSFKAV-RFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEG 415
+ + S F D + + P L ++++ P +VV +D
Sbjct: 566 IESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNC 745
Query: 416 WTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRP---KIIAAV 472
VV+ L+ YS +LESLDA + ++IE ++P KI+ +
Sbjct: 746 ---DRMDVPLPTNVVHVLQCYSALLESLDA---VNVNLDVLQKIERHYIQPTINKIVLSH 907
Query: 473 EAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHER 532
+ PWR F +G P S F + QAECL+ + +RGF V ++ + LVL W +
Sbjct: 908 HNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRK 1087
Query: 533 AMVATSAWRC 542
+++ S WRC
Sbjct: 1088ELISVSTWRC 1117
>TC76809 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 {Vitis
vinifera}, partial (72%)
Length = 2764
Score = 105 bits (261), Expect = 6e-23
Identities = 104/385 (27%), Positives = 171/385 (44%), Gaps = 23/385 (5%)
Frame = +2
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
+ L+ A+ +L+LA+A+++ ++ TG + + A +F AL + G N
Sbjct: 1367 VHTLMACAEAIQQKNLKLAEALVKHISLLASLQTGA-MRKVASYFAQALARRIYG----N 1531
Query: 240 PPRL--SSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLG 297
P SS EI+ F SP F+ FT NQA+LEA G+ +HV+DF + G
Sbjct: 1532 PEETIDSSFSEILHM-----HFYESSPYLKFAHFTANQAILEAFAGAGRVHVIDFGLKQG 1696
Query: 298 IQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRL--IRENLNQFAHDLGIRVQVDFV 355
+Q+ +LM+ +A + G P R+T + P + L + L Q A +G++ +
Sbjct: 1697 MQWPALMQALALRP-GGPPTFRLTGIGPPQADNTDALQQVGWKLAQLAQTIGVQFEFRGF 1873
Query: 356 PLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTA-AFLSDVRRITPGVVVFVDGE 414
+ + + GE A+ + L G+ L+ V++I P +V V+ E
Sbjct: 1874 VCNSIADLDPNMLEIRPGEAVAVNSVFELHTMLARPGSVEKVLNTVKKINPKIVTIVEQE 2053
Query: 415 GWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAG--------GGGEWARRIEM--LLL 464
F +L +YS + +SL+ S ++ G G ++ + M + L
Sbjct: 2054 ANHNGPV---FVDRFTEALHYYSSLFDSLEGSNSSSNNSNSNSTGLGSPSQDLLMSEIYL 2224
Query: 465 RPKI--IAAVEAAGR-----RTTPWREAFYGAGMRPVQLSQFADFQAECLLAK-VQIRGF 516
+I + A E R T WR AG PV L A QA LLA G+
Sbjct: 2225 GKQICNVVAYEGVDRVERHETLTQWRSRMGSAGFEPVHLGSNAFKQASTLLALFAGGDGY 2404
Query: 517 HVAKRQAELVLFWHERAMVATSAWR 541
V + L+L WH R+++ATSAW+
Sbjct: 2405 RVEENNGCLMLGWHTRSLIATSAWK 2479
>TC77063 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 {Vitis
vinifera}, partial (71%)
Length = 1762
Score = 100 bits (248), Expect = 2e-21
Identities = 93/375 (24%), Positives = 160/375 (41%), Gaps = 13/375 (3%)
Frame = +3
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
+ L+ A+ + N+L++A+A+++++ S G + + A +F + L + G +
Sbjct: 603 VHSLMACAEAVEQNNLKMAEALVKQIGYLAVSQEGA-MRKVATYFAEGLARRIYGVFPQH 779
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQ 299
S + +T K F+ FT NQA+LEA G +HV+DF I G+Q
Sbjct: 780 SVSDSLQIHFYETCPNLK----------FAHFTANQAILEAFQGKSSVHVIDFSINQGMQ 929
Query: 300 YASLMKEIAEKAVNGSPLLRITAVVP--EEYAVESRLIRENLNQFAHDLGIRVQVDFVPL 357
+ +LM+ +A + G P R+T + P + + + + L QFA + ++ +
Sbjct: 930 WPALMQALALRP-GGPPAFRLTGIGPPASDNSDHLQQVGWRLAQFAQTIHVQFEYRGFVA 1106
Query: 358 RTFETVSFKAVRFVDGEKTAILLTPAI-FCRLGSEGTA--AFLSDVRRITPGVVVFVDGE 414
+ + + E ++ + +L + A S +R+I P +V V+ E
Sbjct: 1107NSLADLDASMLELRSPETESVAVNSVFELHKLNARPGALEKVFSVIRQIRPEIVTVVEQE 1286
Query: 415 GWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEA 474
A F SL +YS + +SL+ S + + + L +I V
Sbjct: 1287ANHNGPA---FLDRFTESLHYYSTLFDSLEGSSVE----PQDKAMSEVYLGKQICNVVAC 1445
Query: 475 AG-------RRTTPWREAFYGAGMRPVQLSQFADFQAECLLAK-VQIRGFHVAKRQAELV 526
G WR F AG PV L A QA LLA G+ V + L+
Sbjct: 1446EGTDRVERHETLNQWRNRFNSAGFSPVHLGSNAFKQASMLLALFAGGDGYKVEENDGCLM 1625
Query: 527 LFWHERAMVATSAWR 541
L WH R ++ATSAW+
Sbjct: 1626LGWHTRPLIATSAWK 1670
>TC86258 similar to GP|16930433|gb|AAL31902.1 AT4g17230/dl4650c {Arabidopsis
thaliana}, partial (62%)
Length = 2237
Score = 89.7 bits (221), Expect = 3e-18
Identities = 93/396 (23%), Positives = 162/396 (40%), Gaps = 29/396 (7%)
Frame = +3
Query: 176 NWDFIEE----------LIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFK 225
NWD I E L+ A + + A+ ++ + ++ S G P+ R + +
Sbjct: 822 NWDLIAENIPKLDMKEVLLLCAQAVSDGDIPTARGWMDNVLVKMVSVAGDPIQRLSAYLL 1001
Query: 226 DALQSLLSGSNRTNPPRLSSMVEIVQTIRTFK-AFSGISPIPMFSIFTTNQALLEALHGS 284
+ L++ L S L + + T+ I P F+ + N + EA+
Sbjct: 1002 EGLRARLELSGSLIYKSLKCEQPTSKELMTYMHMLYQICPYFKFAYISANAVISEAMANE 1181
Query: 285 LYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESR-----LIRENL 339
+H++DF+I G Q+ L++ +A + G P +RIT V + + +R ++ E L
Sbjct: 1182 SRIHIIDFQIAQGTQWQMLIEALARRP-GGPPFIRITG-VDDSQSFHARGGGLQIVGEQL 1355
Query: 340 NQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTA----- 394
+ FA G+ + + E V + +R GE A+ P + E +
Sbjct: 1356 SNFARSRGVLFEFHSAAMSGCE-VQRENLRVSPGEALAVNF-PFSLHHMPDESVSIENHR 1529
Query: 395 -AFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGG 453
L V+ ++P VV V+ E T + F + V +++FY+ M ES+D VA
Sbjct: 1530 DRLLRLVKSLSPKVVTLVEQESNTN---TSPFFQRFVETMDFYTAMFESID--VACTKDD 1694
Query: 454 EWARRIEMLLLRPKIIAAVEAAGRRTT-------PWREAFYGAGMRPVQLSQFADFQAEC 506
+ +E + I+ + G WR F AG R QLS +
Sbjct: 1695 KKRISVEQNCVARDIVNMIACEGIERVERHEVFGKWRSRFSMAGFRQCQLSSSVMHSVQN 1874
Query: 507 LLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWRC 542
+L + + + R L L W +RAM +SAW C
Sbjct: 1875 MLKDFH-QNYWLEHRDGALYLGWMKRAMATSSAWMC 1979
>TC79190 similar to PIR|E96542|E96542 scarecrow-like protein [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1669
Score = 80.9 bits (198), Expect = 1e-15
Identities = 97/390 (24%), Positives = 163/390 (40%), Gaps = 23/390 (5%)
Frame = +2
Query: 175 SNWDFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSG 234
S D E L A D N ++ + ++ L +++ S +G P+ R + +AL S ++
Sbjct: 404 SRGDLKEMLYTCAKAVDENDIETIEWMVTEL-RKIVSVSGSPIERLGAYMLEALVSKIAS 580
Query: 235 SNRTNPPRLSSMVEIVQTIRTFK-AFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFE 293
S T L + ++ I P F + N A+ EA+ +H++DF+
Sbjct: 581 SGSTIYKSLKCSEPTGNELLSYMHVLYEICPYFKFGYMSANGAIAEAMKEENEVHIIDFQ 760
Query: 294 IGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESR-----LIRENLNQFAHDL-- 346
IG G Q+ SL++ +A + G P +RIT + + Y+ R ++ E L A
Sbjct: 761 IGQGTQWVSLIQALARRP-GGPPKIRITG-IDDSYSSNVRGGGVDIVGEKLLTLAQSCHV 934
Query: 347 -----GIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILL--TPAIFCRLGSEGTAAFLSD 399
+RV V L FE +AV AI+L P + + L
Sbjct: 935 PFEFHAVRVYPSEVRLEDFELRPNEAV----AVNFAIMLHHVPDESVNIHNH-RDRLLRL 1099
Query: 400 VRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRI 459
+ ++P VV V+ E T A F + + ++ +YS + ES+D V + +
Sbjct: 1100AKHMSPKVVTLVEQEFNTN---NAPFLQRFLETMNYYSAVYESID--VVLPRDHKERINV 1264
Query: 460 EMLLLRPKIIAAVEAAGRR-------TTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQ 512
E L +++ V G + WR F AG P LS F + + LL
Sbjct: 1265EQHCLAREVVNLVACEGEERVERHELLSKWRMRFTMAGFTPYPLSSFINSSIKNLLE--S 1438
Query: 513 IRG-FHVAKRQAELVLFWHERAMVATSAWR 541
RG + + +R L L W + ++A+ AWR
Sbjct: 1439YRGHYTLEERDGALFLGWMNQDLIASCAWR 1528
>TC77707 weakly similar to GP|8777405|dbj|BAA96995.1 SCARECROW gene
regulator-like {Arabidopsis thaliana}, partial (65%)
Length = 2161
Score = 72.8 bits (177), Expect = 3e-13
Identities = 82/381 (21%), Positives = 154/381 (39%), Gaps = 18/381 (4%)
Frame = +2
Query: 178 DFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNR 237
D + LI A N+ +L ++E + S G+P+ R + + L + S
Sbjct: 863 DLKQLLIACAKAMAENNTELFDRLIETARNAV-SINGEPIQRLGAYMVEGLVARTEASGN 1039
Query: 238 TNPPRLSSMVEIVQTIRTF-KAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGL 296
+ L + + T+ + I P F N A+ EA ++H++DF+I
Sbjct: 1040 SIYHALKCREPEGEELLTYMQLLFEICPYLKFGYMAANGAIAEACRNEDHIHIIDFQIAQ 1219
Query: 297 GIQYASLMKEIAEKAVNGSPLLRITAV--VPEEYAVESRL--IRENLNQFAHDLGIRVQV 352
G Q+ +L++ +A + G+P +RIT + +YA L + E L+ + GI V+
Sbjct: 1220 GTQWMTLLQALAARP-GGAPHVRITGIDDPVSKYARGKGLEVVGERLSLMSKKFGIPVEF 1396
Query: 353 DFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGT------AAFLSDVRRITPG 406
+P+ V+ + GE A+ P E L V+ ++P
Sbjct: 1397 HGIPV-FGPDVTRDMLDIRHGEALAVNF-PLQLHHTADESVDVNNPRDGLLRLVKSLSPK 1570
Query: 407 VVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRP 466
VV V+ E T F + +L++Y + ES+D +++ + +E L
Sbjct: 1571 VVTLVEQESNTN---TTPFFNRFIETLDYYLAIFESIDVTLSR--NSKERINVEQHCLAR 1735
Query: 467 KIIAAVEAAGRRTT-------PWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVA 519
I+ + G+ W+ AG R LS + + LL + + +
Sbjct: 1736 DIVNVIACEGKERVERHELFGKWKSRLTMAGFRQCPLSSYVNSVIRSLL-RCYSEHYTLV 1912
Query: 520 KRQAELVLFWHERAMVATSAW 540
++ ++L W R +++ SAW
Sbjct: 1913 EKDGAMLLGWKSRNLISASAW 1975
>TC78035 similar to GP|8778540|gb|AAF79548.1| F22G5.9 {Arabidopsis thaliana},
partial (24%)
Length = 2694
Score = 70.5 bits (171), Expect = 2e-12
Identities = 87/379 (22%), Positives = 150/379 (38%), Gaps = 21/379 (5%)
Frame = +3
Query: 183 LIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPR 242
L+ A N + A +L+++ SP+G R A +F +A+++ + G+
Sbjct: 1200 LVLCAQAVSANDNRTANELLKQIRNH-SSPSGDASQRMAHYFANAIEARMVGAGTGTQIL 1376
Query: 243 LSS--MVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQY 300
S M ++ ++ F P F+ F N+ +L+ + +H++DF I G Q+
Sbjct: 1377 YMSQKMFSAADFLKAYQVFISACPFKKFAHFFANKMILKTAEKAETLHIIDFGILYGFQW 1556
Query: 301 ASLMKEIAEKAVNGSPLLRITAV-VPEEYAVESRLIRE---NLNQFAHDLGIRVQVDFVP 356
L+K ++ K G P LRIT + P+ + I E L + + + +P
Sbjct: 1557 PILIKFLS-KVEGGPPKLRITGIEYPQAGFRPAERIEETGRRLANYCERFNVSFEYKAIP 1733
Query: 357 LRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSE------GTAAFLSDVRRITPGVVV- 409
R +ET+ + + E A+ F L E A L +R+I P + V
Sbjct: 1734 SRNWETIQIEDLNIKSNEVVAVNCL-VRFKNLHDETIDVNSPKDAVLKLIRKINPHIFVQ 1910
Query: 410 -FVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKI 468
V+G + A F SL YS M + D ++ EW IE L +I
Sbjct: 1911 SIVNG-----SYNAPFFSTRFKESLFHYSAMFDMYDTLISR--ENEWRSMIEREFLGREI 2069
Query: 469 --IAAVEAAGRRTTP-----WREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKR 521
+ A E R P W+ AG R + L + + L + + F +
Sbjct: 2070 MNVVACEGFERVERPETYKQWQVRNLRAGFRQLPLDKEVMVRFRDKLREWYHKDFVFDED 2249
Query: 522 QAELVLFWHERAMVATSAW 540
++ W R M A++ W
Sbjct: 2250 NNWMLQGWKGRIMYASAGW 2306
>TC82600 similar to GP|9757937|dbj|BAB08425.1 SCARECROW gene regulator-like
protein {Arabidopsis thaliana}, partial (51%)
Length = 1141
Score = 68.9 bits (167), Expect = 5e-12
Identities = 52/170 (30%), Positives = 85/170 (49%), Gaps = 4/170 (2%)
Frame = +2
Query: 183 LIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQS-LLSGSNRTNPP 241
L++ A+C ++L A +L + + L SP G R +F ALQ+ ++S + P
Sbjct: 509 LLQCAECVAMDNLDFANDLLPEITE-LSSPFGTSPERVGAYFAQALQARVVSSCLGSYSP 685
Query: 242 RLSSMVEIVQTIRTFKAF---SGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGI 298
+ V + Q+ R F AF + +SP+ FS FT NQA+ +AL G +H++D +I G+
Sbjct: 686 LTAKSVTLNQSQRIFNAFQSYNSVSPLVKFSHFTANQAIFQALDGEDRVHIIDLDIMQGL 865
Query: 299 QYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRLIRENLNQFAHDLGI 348
Q+ L +A ++ +RIT +ES L FA LG+
Sbjct: 866 QWPGLFHILASRS-KKIRSVRITGFGSSSELLES--TGRRLADFASSLGL 1006
>BM813961 homologue to GP|13365610|db SCARECROW {Pisum sativum}, partial
(21%)
Length = 550
Score = 68.2 bits (165), Expect = 8e-12
Identities = 50/170 (29%), Positives = 83/170 (48%), Gaps = 4/170 (2%)
Frame = -2
Query: 183 LIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGS----NRT 238
L++ A+ +L A +L ++Q L +P G R A +F +A+ + L S T
Sbjct: 543 LLQCAEAVSAENLDQANKMLLEISQ-LSTPFGTSAQRVAAYFSEAISARLVSSCLRIYAT 367
Query: 239 NPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGI 298
PP ++ F+ F+GISP FS FT NQA+ EA +H++D +I G+
Sbjct: 366 LPPHTLHNQKVASA---FQVFNGISPFVKFSHFTANQAIQEAFDREERVHIIDLDIMQGL 196
Query: 299 QYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRLIRENLNQFAHDLGI 348
Q+ L +A + G P +R+T + +E+ + L+ FA LG+
Sbjct: 195 QWPGLFHILASRP-GGPPYVRLTGLGTSMETLEA--TGKRLSDFASKLGL 55
>TC83571 similar to PIR|T10552|T10552 hypothetical protein T12G13.90 -
Arabidopsis thaliana, partial (19%)
Length = 767
Score = 65.1 bits (157), Expect(2) = 2e-11
Identities = 47/144 (32%), Positives = 71/144 (48%), Gaps = 17/144 (11%)
Frame = +1
Query: 197 LAQAILERLNQRL-RSPTGKPLHRAAFHFKDALQSLLSGSN----------------RTN 239
LA+ IL L + + + G + R A HF +AL LL G+ TN
Sbjct: 295 LARVILIPLKELVSQHANGSNMERLAAHFTEALHGLLEGAGGAHNNHHHHNNNKHYLTTN 474
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQ 299
P + T+ F+ +SP F FT NQA++EA+ +HV+D++I G+Q
Sbjct: 475 GPHDNQN----DTLAAFQLLQDMSPYVKFGHFTANQAIIEAVAHERRVHVIDYDIMEGVQ 642
Query: 300 YASLMKEIAEKAVNGSPLLRITAV 323
+ASL++ +A N P LRITA+
Sbjct: 643 WASLIQSLASN--NNGPHLRITAL 708
Score = 21.6 bits (44), Expect(2) = 2e-11
Identities = 8/12 (66%), Positives = 8/12 (66%)
Frame = +3
Query: 165 NNNQPNNNTNSN 176
NNN NNN N N
Sbjct: 165 NNNHNNNNGNKN 200
>TC90344 weakly similar to PIR|H86282|H86282 protein F10B6.34 [imported] -
Arabidopsis thaliana, partial (28%)
Length = 1170
Score = 65.9 bits (159), Expect = 4e-11
Identities = 53/205 (25%), Positives = 94/205 (45%), Gaps = 5/205 (2%)
Frame = +1
Query: 126 QFDQTQ----DFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFIE 181
+FD T DF+ L D+ + ++ ++ N N +S +
Sbjct: 400 EFDHTASLPYDFSQLPDLSAPPPQHHHHQSSSTVN-------------NVEEDSAIKLVH 540
Query: 182 ELIRAADCFDNNHLQLAQAILERLNQRLRSP-TGKPLHRAAFHFKDALQSLLSGSNRTNP 240
L+ AD +L LA +++E + L + T + + A +F DAL + G N
Sbjct: 541 MLMTCADSVQRGNLSLAGSLIEGMQGLLANMNTNSGIGKVAGYFIDALNRRIFGQNN--- 711
Query: 241 PRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQY 300
+S V + + + + P F+ FT NQA+LEA +G +HV+DF + G+Q+
Sbjct: 712 --VSHQVSLYENDVLYHHYYEACPYLKFAHFTANQAILEAFNGHDCVHVIDFNLMHGLQW 885
Query: 301 ASLMKEIAEKAVNGSPLLRITAVVP 325
+L++ +A + G P LR+T + P
Sbjct: 886 PALIQALALRP-GGPPFLRLTGIGP 957
>BI308453 weakly similar to PIR|T02736|T0 probable SCARECROW gene regulator
[imported] - Arabidopsis thaliana, partial (11%)
Length = 712
Score = 60.1 bits (144), Expect = 2e-09
Identities = 43/177 (24%), Positives = 82/177 (46%), Gaps = 4/177 (2%)
Frame = +3
Query: 151 PNTNTNL---DHLVNDFNNNQPNNNTNSNWDFIEELIRAADCFDNNHLQLAQAILERLNQ 207
P NT+ + L N F ++ + D L+ A N + A +L ++ +
Sbjct: 90 PLQNTSFQQNEELSNRFYGSRRQRSYEEVVDLRTLLMLCAQSISCNDISNANQLLNQIKK 269
Query: 208 RLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVE-IVQTIRTFKAFSGISPIP 266
SPTG R A F +AL++ L+G+ LSS + ++ ++ +S P
Sbjct: 270 H-SSPTGDGTQRLAHFFGNALEARLAGTGSHVYRALSSKKKSAADMVKAYQVYSSACPFE 446
Query: 267 MFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAV 323
+I +N A+L + +H++DF +G G ++ + +++++ G P LRIT +
Sbjct: 447 KLAIMFSNDAILNVAKETESLHIIDFGVGYGFKWLGFIYRLSKRS-GGPPKLRITGI 614
>TC77896 weakly similar to GP|22087131|gb|AAM90848.1 hairy meristem {Petunia
x hybrida}, partial (8%)
Length = 258
Score = 57.0 bits (136), Expect = 2e-08
Identities = 32/81 (39%), Positives = 48/81 (58%)
Frame = +3
Query: 219 RAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALL 278
RA + K+ALQ LL + + S ++ I +K+FS ISP+ F+ FT+NQALL
Sbjct: 15 RAGCYCKEALQLLLQSNVNNSNNNSFSGTRLLLKIGAYKSFSQISPVLQFANFTSNQALL 194
Query: 279 EALHGSLYMHVVDFEIGLGIQ 299
EA+ +H++D +IG G Q
Sbjct: 195 EAVERFDRIHIIDCDIGFGGQ 257
>TC77716 similar to GP|20465587|gb|AAM20276.1 unknown protein {Arabidopsis
thaliana}, partial (63%)
Length = 2392
Score = 57.0 bits (136), Expect = 2e-08
Identities = 84/387 (21%), Positives = 149/387 (37%), Gaps = 28/387 (7%)
Frame = +3
Query: 183 LIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRT---- 238
L A + A A+++ L Q L S G+P R A + + L + L+ S +
Sbjct: 903 LYECASALSEGNEVKASAMIDDLRQ-LVSIQGEPSDRIAAYMVEGLAARLASSGKCIYKA 1079
Query: 239 ----NPP---RLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVD 291
PP RL++M + + FK F N A+ EA+ +H++D
Sbjct: 1080 LKCKEPPSSDRLAAMQILFEVCPCFK----------FGFIAANGAISEAIKDDKKVHIID 1229
Query: 292 FEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVES----RLIRENLNQFAHDLG 347
F+I G QY +L++ +A + P +R+T V E S +I + L + A LG
Sbjct: 1230 FDINQGSQYITLIQTLASRP-GKPPYVRLTGVDDPESVQRSVGGLSIIGQRLEKLAEVLG 1406
Query: 348 IRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAA------FLSDVR 401
+ + V R+ V+ + GE + + + E + L V+
Sbjct: 1407 LPFEFRAVASRS-SIVTPSMLNCRPGEALVVNFAFQLH-HMPDESVSTVNERDQLLRMVK 1580
Query: 402 RITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEM 461
+ P +V V+ + T + F V + +YS + ESLDA++ + +E
Sbjct: 1581 SLNPKLVTVVEQDVNTN---TSPFLPRFVEAYNYYSAVFESLDATLPR--ESQDRVNVER 1745
Query: 462 LLLRPKIIAAVEAAGRR-------TTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIR 514
L I+ + G WR AG +S +A L +
Sbjct: 1746 QCLARDIVNVIACEGEDRIERYEVAGKWRARMKMAGFTSSPMSTNVK-EAIRELIRQYCD 1922
Query: 515 GFHVAKRQAELVLFWHERAMVATSAWR 541
+ + + L W ++ ++ SAW+
Sbjct: 1923 KYKIIEEMGGLHFGWEDKNLIVASAWK 2003
>TC80896 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 {Vitis
vinifera}, partial (17%)
Length = 783
Score = 55.8 bits (133), Expect = 4e-08
Identities = 45/157 (28%), Positives = 66/157 (41%), Gaps = 7/157 (4%)
Frame = +1
Query: 392 GTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGG 451
G L +R + P +V V+ E F +L +YS + +SL+A
Sbjct: 16 GIEMVLGWIRSLNPKIVTVVEQEANHNENG---FMERFTEALHYYSTVFDSLEACPV--- 177
Query: 452 GGEWARRIEMLLLRPKIIAAVEAAG----RRTTP---WREAFYGAGMRPVQLSQFADFQA 504
E + + + L+ +I V G R P W+E AG RP+ L A QA
Sbjct: 178 --EPDKAMAEMYLQREICNVVCCEGPARVERHEPLVKWKERLGKAGFRPLHLGSNAFKQA 351
Query: 505 ECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
LL G+ V + Q L L WH R ++A SAW+
Sbjct: 352 SMLLTLFSAEGYCVEENQGSLTLGWHSRPLIAASAWQ 462
>TC80602 similar to GP|14517552|gb|AAK62666.1 F17J6.12/F17J6.12 {Arabidopsis
thaliana}, partial (31%)
Length = 1190
Score = 54.7 bits (130), Expect = 9e-08
Identities = 38/142 (26%), Positives = 67/142 (46%), Gaps = 1/142 (0%)
Frame = +2
Query: 183 LIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPR 242
L A +N L +AQ +++ L Q + S +G+P+ R + + L + LS S +
Sbjct: 737 LTACAKAIADNDLLMAQWLMDELRQMV-SVSGEPIQRLGAYMLEGLVARLSASGSSIYKS 913
Query: 243 LSSMV-EIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYA 301
L E + + + P F + N A+ EA+ +H++DF+I G Q+
Sbjct: 914 LRCKEPESAELLSYMNILYEVCPYFKFGYMSANGAIAEAMKNEARVHIIDFQIAQGSQWI 1093
Query: 302 SLMKEIAEKAVNGSPLLRITAV 323
SL++ A + G P +RIT +
Sbjct: 1094SLIQAFAARP-GGPPHIRITGI 1156
>BG587215 similar to GP|8453100|gb| short-root protein {Arabidopsis
thaliana}, partial (31%)
Length = 651
Score = 49.7 bits (117), Expect = 3e-06
Identities = 53/200 (26%), Positives = 83/200 (41%), Gaps = 17/200 (8%)
Frame = +3
Query: 141 SNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNS---NWDFIEELIRAADCFDNNHLQL 197
S+ YN P T+ N N P + DF EL +N L+
Sbjct: 81 SSKHYYPYNQPQTS-------NTITTNTPTTSLTPPPPTADFTFEL--TGRWANNILLET 233
Query: 198 AQAILERLNQRLR----------SPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMV 247
A+AI E+ + RL+ SP G + A +F AL S ++ + L+S
Sbjct: 234 ARAIAEKNSTRLQQLMWMLNELSSPYGDIEQKLAAYFLQALFSRMTEAGTRTFRTLASAS 413
Query: 248 E----IVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASL 303
E T +T F +SP F N A+LEA G +H++D Q+ +L
Sbjct: 414 EKTCSFESTRKTVLKFQEVSPWTTFGHVACNGAILEAFEGDSKLHIIDISNTYCTQWPTL 593
Query: 304 MKEIAEKAVNGSPLLRITAV 323
+ +A +A + +P LR+T +
Sbjct: 594 FEALATRA-DDTPHLRLTTI 650
Score = 28.1 bits (61), Expect = 9.0
Identities = 16/42 (38%), Positives = 21/42 (49%), Gaps = 3/42 (7%)
Frame = +3
Query: 8 VSSSSSS---PQPKLTTTQNIVVNVPTPNPTPTPTTTTLCYE 46
+SSSSS P + T+ I N PT + TP P T +E
Sbjct: 69 LSSSSSKHYYPYNQPQTSNTITTNTPTTSLTPPPPTADFTFE 194
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,365,110
Number of Sequences: 36976
Number of extensions: 271069
Number of successful extensions: 3255
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 2538
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2947
length of query: 542
length of database: 9,014,727
effective HSP length: 101
effective length of query: 441
effective length of database: 5,280,151
effective search space: 2328546591
effective search space used: 2328546591
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148484.4


