

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.2 - phase: 0
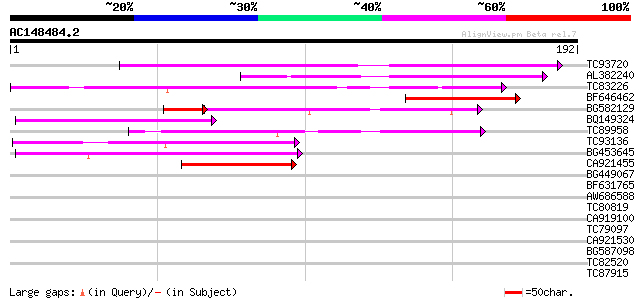
(192 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93720 similar to GP|21743320|dbj|BAC03314. putative nematode r... 80 5e-16
AL382240 69 9e-13
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 62 1e-10
BF646462 50 4e-07
BG582129 40 8e-07
BQ149324 48 2e-06
TC89958 similar to GP|12483623|dbj|BAB21445. NADH dehydrogenase ... 45 2e-05
TC93136 44 3e-05
BG453645 40 4e-04
CA921455 40 7e-04
BG449067 35 0.014
BF631765 33 0.068
AW686588 33 0.068
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 28 0.10
CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [i... 32 0.20
TC79097 weakly similar to GP|20197587|gb|AAD14519.2 expressed pr... 31 0.26
CA921530 31 0.34
BG587098 30 0.75
TC82520 29 0.99
TC87915 similar to SP|O67750|FLIP_AQUAE Flagellar biosynthetic p... 28 2.2
>TC93720 similar to GP|21743320|dbj|BAC03314. putative nematode
resistance-like protein {Oryza sativa (japonica
cultivar-group)}, partial (3%)
Length = 457
Score = 80.1 bits (196), Expect = 5e-16
Identities = 44/150 (29%), Positives = 70/150 (46%)
Frame = -2
Query: 38 VFALLDMLTTKQRVVFAATVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKT 97
+F LL ++ Q + A +WSLWK +K+W+ E + VV A ++ +W+ A T
Sbjct: 438 IFTLLQQFSSDQCELMATVMWSLWKSCRMKLWQQNNESNSQVVHHATHLLNDWRAAQTI* 259
Query: 98 PAESRAATTAPPTQLLSNSAEVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGIC 157
+ P + + W T YKCN+DA F ++ N+ G+G+C
Sbjct: 258 SHNHDQSNMVYPCPMKQDEEA----------WMTLTPDMYKCNIDASFFTSLNKVGLGMC 109
Query: 158 IHDEEGAFVLAKMTSFPCLHQVTVGEAMGL 187
+ D+ FVLAK F + GEA+ L
Sbjct: 108 LADDADDFVLAKTIWFAPSCDIDAGEAVRL 19
>AL382240
Length = 320
Score = 69.3 bits (168), Expect = 9e-13
Identities = 38/104 (36%), Positives = 54/104 (51%)
Frame = +3
Query: 79 VVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEVNSSDDGLVLWQKPTSGRYK 138
V RA + EW+ + + S +T P Q +V W+KP G YK
Sbjct: 9 VFQRATHVFEEWRTTQ-RIRSSSNTQSTTPQVQPSRPEEDV---------WKKPAPGXYK 158
Query: 139 CNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSFPCLHQVTVG 182
CN+DA FS++FN G+G+C+ DE+ AFVLA+ F L + VG
Sbjct: 159 CNIDASFSTSFNIVGLGMCLRDEDDAFVLARTEWFAPLCDIDVG 290
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 62.4 bits (150), Expect = 1e-10
Identities = 49/171 (28%), Positives = 78/171 (44%), Gaps = 3/171 (1%)
Frame = +3
Query: 1 MAHLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDMLTTKQRVV---FAATV 57
+ HLF +C + +VW S L N ++ N + L + + K + AA +
Sbjct: 273 ITHLFMSCPLSKRVWFGSNL-----CINFDNLPNPNFIN*LYEAIL*KDECITI*IAAII 437
Query: 58 WSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSA 117
++LW RNL V ED T + ++ RA I++++ ANT+ P P +Q +
Sbjct: 438 YNLWHARNLSVLEDQTILEMDIIQRASNCISDYKQANTQAPPSMARTGYDPRSQ---HRP 608
Query: 118 EVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLA 168
N+ W++P G K N DA N + G+GI I DE G + A
Sbjct: 609 AKNTK------WKRPNLGLVKVNTDANL-QNHGKWGLGIIIRDEVGLVMAA 740
>BF646462
Length = 487
Score = 50.4 bits (119), Expect = 4e-07
Identities = 22/39 (56%), Positives = 25/39 (63%)
Frame = -1
Query: 135 GRYKCNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSF 173
GR KCN+D F + N TGI CI DEEG F LAK+ F
Sbjct: 355 GRVKCNIDXAFFEH*NNTGISFCIRDEEGTFXLAKIMPF 239
>BG582129
Length = 534
Score = 40.0 bits (92), Expect(2) = 8e-07
Identities = 29/107 (27%), Positives = 47/107 (43%), Gaps = 13/107 (12%)
Frame = -3
Query: 67 KVWEDVTEVAAVVVDRARVMITEWQIANTKTPAE----SRAATTAPPTQLLSNSAEVNSS 122
K+W+ VT+ + + + A ++ W +AN K R+ + SN+++
Sbjct: 454 KIWQLVTKTNSQI*ECANHVLE*WMLANIKKDFRLKVIMRSVQWSGAGAATSNTSKTRQQ 275
Query: 123 DDGLVLWQKPTSGRYKCNVDAVFSSN---------FNRTGIGICIHD 160
L WQ P GR K NVDA FS++ F G+C H+
Sbjct: 274 ---LCTWQPPPVGRLKYNVDASFSASSTK*V*ECVFGMMRAGLCQHE 143
Score = 34.7 bits (78), Expect = 0.023
Identities = 13/21 (61%), Positives = 17/21 (80%)
Frame = -2
Query: 149 FNRTGIGICIHDEEGAFVLAK 169
FN+ GIG+CI D+EG FV A+
Sbjct: 206 FNKVGIGVCIRDDEGRFVSAR 144
Score = 28.9 bits (63), Expect(2) = 8e-07
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = -1
Query: 53 FAATVWSLWKHRNLK 67
FA +W +WKHRNL+
Sbjct: 495 FATILWRIWKHRNLR 451
>BQ149324
Length = 374
Score = 48.1 bits (113), Expect = 2e-06
Identities = 22/68 (32%), Positives = 34/68 (49%)
Frame = +1
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDMLTTKQRVVFAATVWSLWK 62
HL F CS ++ VW + S +S D + +F L L+ + +F +WS+ K
Sbjct: 91 HLIFQCSSSLNVWSMLPFLSTISILLQQDMDSKNIIFKALHDLSNEDAALFCCVLWSI*K 270
Query: 63 HRNLKVWE 70
RN KVW+
Sbjct: 271 QRNNKVWK 294
>TC89958 similar to GP|12483623|dbj|BAB21445. NADH dehydrogenase subunit 4L
{Gonostoma gracile}, partial (17%)
Length = 1040
Score = 45.1 bits (105), Expect = 2e-05
Identities = 37/122 (30%), Positives = 50/122 (40%), Gaps = 1/122 (0%)
Frame = +2
Query: 41 LLDMLTTKQRVVFAATVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITE-WQIANTKTPA 99
LLD+L + A W + K RN K+W V E +V AR + W N+
Sbjct: 329 LLDLLR-----LLATISWRIKKMRNRKLW*GV*ETTDQIVQCARNQYMQSWLYVNSVQTT 493
Query: 100 ESRAATTAPPTQLLSNSAEVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICIH 159
AP SA V +V W+ P+ G KCN+DA +N G G +
Sbjct: 494 N----VVAPEQVQQQTSAAV------MVPWEPPS*GYVKCNMDAALFKEYNCVGAGFRV* 643
Query: 160 DE 161
DE
Sbjct: 644 DE 649
>TC93136
Length = 722
Score = 44.3 bits (103), Expect = 3e-05
Identities = 25/99 (25%), Positives = 46/99 (46%), Gaps = 2/99 (2%)
Frame = +1
Query: 2 AHLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDMLTTKQRV--VFAATVWS 59
+HLF C F VW W S VF LL+ + + V ++ AT+W
Sbjct: 271 SHLFLHCPFLSSVWSKILGWLYYS--------KFVCVFRLLERRSRTKGVWLIWHATIWV 426
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTP 98
+WK N ++++++++ +V+ +V+ W + K P
Sbjct: 427 IWKGINNRIFKNISKAIDEIVEEIKVLSWRWSLTRLKCP 543
>BG453645
Length = 622
Score = 40.4 bits (93), Expect = 4e-04
Identities = 25/100 (25%), Positives = 44/100 (44%), Gaps = 3/100 (3%)
Frame = +3
Query: 3 HLFFACSFAIQVWRLSGLWSDVS---AANSNDDSNIGTVFALLDMLTTKQRVVFAATVWS 59
H+F C A VW W DV+ N + + +T +V+ T+W
Sbjct: 69 HIFLHCDVASLVWFRVMKWFDVNFLIPPNLFVHWECWSEGGSANRVTKGHWLVWHTTIWV 248
Query: 60 LWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPA 99
LW RN +++ + VA V++ +V+ W + + TP+
Sbjct: 249 LWAKRNDLIFKGLNCVAEDVIEEIKVLS*RWMLERSSTPS 368
>CA921455
Length = 713
Score = 39.7 bits (91), Expect = 7e-04
Identities = 16/39 (41%), Positives = 25/39 (64%)
Frame = -3
Query: 59 SLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKT 97
SLW+ RN K+WE + E V+ RA+ + + WQ A+ +T
Sbjct: 648 SLWRRRNTKLWESILEQCLHVMARAKEVFSLWQFAHKET 532
Score = 28.9 bits (63), Expect = 1.3
Identities = 15/52 (28%), Positives = 22/52 (41%)
Frame = -1
Query: 139 CNVDAVFSSNFNRTGIGICIHDEEGAFVLAKMTSFPCLHQVTVGEAMGLFEA 190
CN+D S + + +C+ D E F C+ GEA GL +A
Sbjct: 464 CNLDVAIFSTMKKVSMTVCLRDVE*HFKKTYNIWCTCVMSAAEGEA*GLNQA 309
>BG449067
Length = 578
Score = 35.4 bits (80), Expect = 0.014
Identities = 31/93 (33%), Positives = 43/93 (45%), Gaps = 4/93 (4%)
Frame = -2
Query: 102 RAATTAPPTQL---LSNSAEVNSSDDGLVLWQKPTSGRYKCNVDAVFSSNFNRTGIGICI 158
R A A P + L +S + S V W+KP K N DA SS + GIG+
Sbjct: 397 RKAQLADPNSIDSSLPSSTQHRVSVPKHVKWKKPEKDIIKLNSDANLSST-DLWGIGVVA 221
Query: 159 HDEEGAFVLAKMTSFPC-LHQVTVGEAMGLFEA 190
++EG F +A T F T EA G+++A
Sbjct: 220 RNDEG-FAMASGTWFRFGFPSATTAEAWGIYQA 125
>BF631765
Length = 498
Score = 33.1 bits (74), Expect = 0.068
Identities = 18/49 (36%), Positives = 29/49 (58%), Gaps = 2/49 (4%)
Frame = +1
Query: 48 KQRV--VFAATVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIAN 94
+QRV +F ++W +WKHRN KV+ +V +V ++ E+ IAN
Sbjct: 97 EQRVKQLFGTSLWLIWKHRNQKVFNNVDFDPPLV----SALVEEFSIAN 231
>AW686588
Length = 567
Score = 33.1 bits (74), Expect = 0.068
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 4/71 (5%)
Frame = +1
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDMLTTKQR----VVFAATVW 58
HLF C+ VW+ W VS A+ +D S+ F T +R +++ VW
Sbjct: 283 HLFLHCATFGAVWQHIRAWIGVSGADPHDLSDHFIQFITCTGHTRARRSFMQLIWLLCVW 462
Query: 59 SLWKHRNLKVW 69
+W RN +++
Sbjct: 463 MVWNERNNRLF 495
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 28.5 bits (62), Expect(2) = 0.10
Identities = 9/39 (23%), Positives = 20/39 (51%)
Frame = +1
Query: 52 VFAATVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEW 90
+ W +WK RN +++++ V++DR ++ W
Sbjct: 928 ILQVNFWVIWKERNNRLFQNTVTAPFVLIDRIKLHSFLW 1044
Score = 22.7 bits (47), Expect(2) = 0.10
Identities = 8/29 (27%), Positives = 14/29 (47%)
Frame = +2
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSND 31
HLF C+ +W L W + + +S +
Sbjct: 782 HLFLHCNVFCSLWSLVRNWLGIPSMSSGE 868
>CA919100 homologue to PIR|G90291|G902 endoglucanase precursor [imported] -
Sulfolobus solfataricus, partial (3%)
Length = 789
Score = 31.6 bits (70), Expect = 0.20
Identities = 19/92 (20%), Positives = 38/92 (40%), Gaps = 4/92 (4%)
Frame = -2
Query: 3 HLFFACSFAIQVWRLSGLWSDVSAANSN--DDSNIGTVFALLDMLTTKQ--RVVFAATVW 58
HLF +CS+ +W L W ++N D + V + ++ ++++ W
Sbjct: 428 HLFLSCSYFASLWSLVRDWIGFVGVDTNVLSDHFVQFVHSTGGNKASQSFLQLIWLLCAW 249
Query: 59 SLWKHRNLKVWEDVTEVAAVVVDRARVMITEW 90
LW RN + D ++D+ + + W
Sbjct: 248 VLWTERNNMCFNDSITPLPRLLDKVKYLSLGW 153
>TC79097 weakly similar to GP|20197587|gb|AAD14519.2 expressed protein
{Arabidopsis thaliana}, partial (3%)
Length = 1539
Score = 31.2 bits (69), Expect = 0.26
Identities = 21/52 (40%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Frame = +3
Query: 138 KCNVDAVFSSNF--NRTGIGICIHDEEGAFVLAKMTSFPCLHQVTVGEAMGL 187
KC+VDA + N GIGICI+++ G F+ AK L EAM L
Sbjct: 600 KCSVDAGAPLFYQQNMYGIGICINNDSGEFI*AKTKHKKQLLSPKEAEAMRL 755
>CA921530
Length = 793
Score = 30.8 bits (68), Expect = 0.34
Identities = 11/48 (22%), Positives = 24/48 (49%)
Frame = -1
Query: 51 VVFAATVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTP 98
+++ AT+W LWK N +++ + A +++ +V+ W P
Sbjct: 595 LIWHATIWILWKAWNNRIFNNQAVEAEDIIEEVKVLSWRWTFNRINIP 452
>BG587098
Length = 402
Score = 29.6 bits (65), Expect = 0.75
Identities = 36/152 (23%), Positives = 56/152 (36%), Gaps = 8/152 (5%)
Frame = +2
Query: 10 FAIQVWRLSGLWSDVSAANSNDDSNIGTVFALLDMLTTK-------QRVVFAATVWSLWK 62
FA +VW+L L V A ++I AL++ TT V +W++W
Sbjct: 11 FAQEVWKLIPLRQVVHLA-----TDINFKQALVEFRTTVCFPPSGIAMTVLPWVLWAIWS 175
Query: 63 HRNLKVWEDVTEVAAVVVDRARVMITEWQIANTKTPAESRAATTAPPTQLLSNSAEVNSS 122
RNL V+E+ +A + EW A + + + T+ + N+ E
Sbjct: 176 TRNLHVFENWILSPMETTTKAPNLGREWNNAQQQIQSVKKVIPTS--RRSTGNTGE---- 337
Query: 123 DDGLVLWQKPTSGRY-KCNVDAVFSSNFNRTG 153
SG Y C DA + R G
Sbjct: 338 -----------SGAYTTCRSDAAYDKRSKRAG 400
>TC82520
Length = 833
Score = 29.3 bits (64), Expect = 0.99
Identities = 11/41 (26%), Positives = 25/41 (60%)
Frame = +2
Query: 50 RVVFAATVWSLWKHRNLKVWEDVTEVAAVVVDRARVMITEW 90
++++ A+VW LWK RN +V+++ + V++ ++ W
Sbjct: 494 QIMWFASVWVLWKKRNNRVFQNSLSDPSTFVEQVKMHSFLW 616
>TC87915 similar to SP|O67750|FLIP_AQUAE Flagellar biosynthetic protein
fliP. {Aquifex aeolicus}, partial (7%)
Length = 976
Score = 28.1 bits (61), Expect = 2.2
Identities = 17/59 (28%), Positives = 25/59 (41%)
Frame = -2
Query: 86 MITEWQIANTKTPAESRAATTAPPTQLLSNSAEVNSSDDGLVLWQKPTSGRYKCNVDAV 144
+IT+ + + T S + PP L S +A SS + PTSG C D +
Sbjct: 855 LITQTKRLHFLTKTTS*LISPLPPKYLYSGNASAVSSPSSCFIMTDPTSGILSC*ADLI 679
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.130 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,890,077
Number of Sequences: 36976
Number of extensions: 89225
Number of successful extensions: 471
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 464
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 466
length of query: 192
length of database: 9,014,727
effective HSP length: 91
effective length of query: 101
effective length of database: 5,649,911
effective search space: 570641011
effective search space used: 570641011
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148484.2


