

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
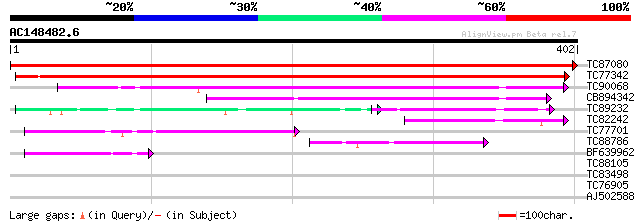
Query= AC148482.6 - phase: 0
(402 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87080 similar to GP|13785209|emb|CAC37356. putative membrane p... 814 0.0
TC77342 similar to GP|13785211|emb|CAC37357. putative membrane p... 543 e-155
TC90068 similar to GP|20197988|gb|AAD22321.2 expressed protein {... 214 6e-56
CB894342 similar to GP|20197988|gb expressed protein {Arabidopsi... 186 1e-47
TC89232 similar to PIR|T47967|T47967 hypothetical protein F15G16... 69 2e-24
TC82242 weakly similar to GP|21592781|gb|AAM64730.1 putative mem... 90 2e-18
TC77701 similar to PIR|T47789|T47789 hypothetical protein F17J16... 89 3e-18
TC88786 weakly similar to GP|7268619|emb|CAB78828.1 unknown prot... 63 2e-10
BF639962 weakly similar to GP|21593814|gb| unknown {Arabidopsis ... 60 1e-09
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 31 0.97
TC83498 similar to GP|10120315|emb|CAC08184. beta-ketoacyl-acyl ... 30 2.2
TC76905 similar to PIR|T48939|T48939 acidic ribosomal protein P2... 29 3.7
AJ502588 similar to GP|7268619|emb| unknown protein {Arabidopsis... 29 3.7
>TC87080 similar to GP|13785209|emb|CAC37356. putative membrane protein
{Solanum tuberosum}, partial (47%)
Length = 1483
Score = 814 bits (2102), Expect = 0.0
Identities = 402/402 (100%), Positives = 402/402 (100%)
Frame = +1
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 60
MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT
Sbjct: 55 MVKNLRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQT 234
Query: 61 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 120
TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS
Sbjct: 235 TGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANS 414
Query: 121 RTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMT 180
RTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMT
Sbjct: 415 RTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMT 594
Query: 181 SANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKS 240
SANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKS
Sbjct: 595 SANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKS 774
Query: 241 ADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFAL 300
ADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFAL
Sbjct: 775 ADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFAL 954
Query: 301 LLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIA 360
LLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIA
Sbjct: 955 LLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIA 1134
Query: 361 ALGGVAVLLEAYTWIIVIKRKKSENKLQGMNGTNGNGYGSRV 402
ALGGVAVLLEAYTWIIVIKRKKSENKLQGMNGTNGNGYGSRV
Sbjct: 1135ALGGVAVLLEAYTWIIVIKRKKSENKLQGMNGTNGNGYGSRV 1260
>TC77342 similar to GP|13785211|emb|CAC37357. putative membrane protein
{Solanum tuberosum}, partial (47%)
Length = 1581
Score = 543 bits (1398), Expect = e-155
Identities = 266/395 (67%), Positives = 316/395 (79%), Gaps = 2/395 (0%)
Frame = +2
Query: 5 LRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKL 64
LR + +L + T + TQ+CK+Q F++N FT+CRDLPQLTSYLHW+YD+T+GKL
Sbjct: 116 LRLTLALCVLSSIILT-SSAQTQSCKNQTFSDNRAFTTCRDLPQLTSYLHWSYDETSGKL 292
Query: 65 DIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQL 124
DIA+ H GIT TNRWVAWAINP+ +L +M GAQALVAI Q+SG+PKAYTS+I ++ T+L
Sbjct: 293 DIAYIHTGITATNRWVAWAINPSRNLDPAMIGAQALVAIPQASGSPKAYTSNIIDTSTRL 472
Query: 125 AESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSG-STPQMHDMTSAN 183
E ISYP SGL AT++NNEVTI+A++TLP GT S VH+WQDG +S STPQ H S++
Sbjct: 473 QEGTISYPVSGLSATYQNNEVTIFATLTLPNGTTSFVHVWQDGVLSSDSTPQEHSHESSH 652
Query: 184 TQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADP 243
SKE LDL SG S+ SG GS RRRNTHGVLNAISWGILMP GAVIARYLKVFKSADP
Sbjct: 653 QNSKEVLDLVSGTSQAASGIGSRQRRRNTHGVLNAISWGILMPTGAVIARYLKVFKSADP 832
Query: 244 AWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLR 303
AWFYLH+TCQ +AYIVG++G+GTGLKLGSDS G+TY THR L IV+ L TLQVFAL LR
Sbjct: 833 AWFYLHITCQVSAYIVGLSGFGTGLKLGSDSVGITYDTHRALAIVLVTLATLQVFALFLR 1012
Query: 304 PKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALG 363
P KDHK+RFYWN+YH VGY TI ISI+N+FKGFEAL DRY NWKHAY GII ALG
Sbjct: 1013PNKDHKLRFYWNIYHHVVGYVTISISIVNVFKGFEALGDFVGDRYKNWKHAYIGIIGALG 1192
Query: 364 GVAVLLEAYTWIIVIKRKKSENKL-QGMNGTNGNG 397
G+AVLLEAYTW++ +KRKK+ENK G+NG NG+G
Sbjct: 1193GIAVLLEAYTWMVCMKRKKAENKTSDGVNGANGHG 1297
>TC90068 similar to GP|20197988|gb|AAD22321.2 expressed protein {Arabidopsis
thaliana}, partial (88%)
Length = 1352
Score = 214 bits (544), Expect = 6e-56
Identities = 127/379 (33%), Positives = 197/379 (51%), Gaps = 17/379 (4%)
Frame = +1
Query: 35 TNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKGITDTNRWVAWAINPNNDLASSM 94
T+ F C +LP + + WT + L++ F I+ + WV + INP + M
Sbjct: 73 TSTKTFQKCMNLPTQQASIAWTLHPHSSTLELIFFGTFISPSG-WVGFGINPTSP---QM 240
Query: 95 NGAQALVAILQSSGTPKAYTSSIANSRTQLAESNI-SYP-------------HSGLIAT- 139
G AL++ + I ++ +L +S + S P + G +AT
Sbjct: 241 TGTNALISFPDPNTGQIVLLPFILDTTVKLQKSPLLSQPFDNINLLSSSAAMYGGKMATI 420
Query: 140 HENNEVTIYASITLPVGTPSLVHLWQDGA-MSGSTPQMHDMTSANTQSKESLDLRSGASE 198
H + IYA + L + +W G + G +P +H TS + S + D+ SG+S
Sbjct: 421 HNGAPIQIYAKLKLESNKTKIHLVWNRGLYVQGYSPTIHPTTSIDLSSIATFDVLSGSSS 600
Query: 199 QGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYI 258
S L+ R HGVLNAISWGIL+P GA+ ARYL+ F++ P+WFY H Q +I
Sbjct: 601 SSSQHTDLTMLRVIHGVLNAISWGILLPTGAITARYLRHFQTLGPSWFYAHAGIQMFGFI 780
Query: 259 VGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYH 318
+G G+G G++L + GV Y HR LGI +FCLG LQ ALL RP +K R YW YH
Sbjct: 781 LGTVGFGIGIQLRKMTPGVEYGLHRKLGIAVFCLGALQTLALLFRPNTTNKFRKYWKSYH 960
Query: 319 WGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWIIVI 378
VGY+ +++ +N+F+GFE + S + K +Y ++ L GV++ LE +W+I
Sbjct: 961 HFVGYSCVVLGFVNVFQGFEVIGASRSYA----KLSYCLGLSTLIGVSIALEVNSWVIFC 1128
Query: 379 KRKKSEN-KLQGMNGTNGN 396
++ K E + +G+ GT+ +
Sbjct: 1129RKSKEEKMRKEGLIGTSSD 1185
>CB894342 similar to GP|20197988|gb expressed protein {Arabidopsis thaliana},
partial (64%)
Length = 838
Score = 186 bits (473), Expect = 1e-47
Identities = 97/246 (39%), Positives = 140/246 (56%), Gaps = 1/246 (0%)
Frame = +3
Query: 140 HENNEVTIYASITLPVGTPSLVHLWQDGA-MSGSTPQMHDMTSANTQSKESLDLRSGASE 198
H + IYA L + +W G + G +P +H TS + S +LD+ SG+S
Sbjct: 108 HNGAPIQIYAKFKLESNKTKIHLVWNRGLYVQGYSPTIHPTTSTDLSSIATLDVLSGSSA 287
Query: 199 QGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYI 258
+ L+ R HG LNAISWGIL+P+GA+ ARY + +S PAWFY H Q A+I
Sbjct: 288 RQHT--DLTMLRVIHGTLNAISWGILLPMGAITARYFRHIQSLGPAWFYAHAGIQLFAFI 461
Query: 259 VGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYH 318
+G G+ G+ LG S GV YS HR LG+ +FCLG LQ ALL RP +K R YW YH
Sbjct: 462 LGTVGFAIGIHLGQLSPGVEYSLHRKLGVAVFCLGALQTLALLFRPNARNKFRKYWKSYH 641
Query: 319 WGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWIIVI 378
VGY+ +++ +N+F+GFE + S + K +Y ++ L GV++ LE +W++
Sbjct: 642 HFVGYSCVVLGFVNVFQGFEVMGASRSYA----KLSYCLGLSTLIGVSIALEVNSWVMFC 809
Query: 379 KRKKSE 384
++ K E
Sbjct: 810 RKSKEE 827
>TC89232 similar to PIR|T47967|T47967 hypothetical protein F15G16.140 -
Arabidopsis thaliana, partial (38%)
Length = 1224
Score = 68.6 bits (166), Expect(2) = 2e-24
Identities = 69/277 (24%), Positives = 111/277 (39%), Gaps = 17/277 (6%)
Frame = +3
Query: 5 LRFMFLISILMFVTTTLAQTTTQ---TCKSQNFT-------NNAIFTSCRDLPQLTSYLH 54
+R L I F T L T TCK+ NF N C+ + T L
Sbjct: 60 VRICMLNLIFNFKTIALGDDTASSESTCKNTNFQIFLPPPYQNISSPICKPVWH-TYELR 236
Query: 55 WTYDQTTGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYT 114
+T + T + ++ + T WV + + M G+ A+V + G K
Sbjct: 237 YTKNGDTTTIILSAPY-----TVGWVGIGFSRDG----KMVGSSAMVGWINKHGHAKIKQ 389
Query: 115 SSI-ANSRTQLAESNISYPHSGLIATHENNEVTIYASI----TLPVGTPSLVHLWQDGAM 169
+ N +++ + P + + A N IY + T+P G ++ A
Sbjct: 390 YYLRGNKSSEVIANKGELPLNNVPAAVATNGAEIYLAFQLQTTIPFGKQPILL-----AF 554
Query: 170 SGSTPQMHDMTSANTQSKESLDLRSGASE--QGSGGGSLSRRRNTHGVLNAISWGILMPL 227
S P H ++ + ++ D SGA SL + R HG++ I WG+++P+
Sbjct: 555 STKHPHEHHLSKHDDKTTIIFDFSSGAKSGSMDPASNSLIQMRKNHGIIGIIGWGLILPV 734
Query: 228 GAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGW 264
GA++ARY FK + WFYLH S VG+ W
Sbjct: 735 GAIVARY---FKHKETLWFYLH----SIIQFVGICIW 824
Score = 61.6 bits (148), Expect(2) = 2e-24
Identities = 41/130 (31%), Positives = 62/130 (47%)
Frame = +1
Query: 257 YIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNL 316
+ G+A GL+L S V HR +GI + L LQ+ A LRP +D K R WNL
Sbjct: 814 FAFGLATVLLGLQLYSQMH-VHIPAHRGIGIFVLVLSILQILAFFLRPNRDSKFRKMWNL 990
Query: 317 YHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWII 376
YH G + + +NI G +A + A WK +Y + + V ++LE ++
Sbjct: 991 YHSWFGRMALFFAALNIVLGMQAAGATNA-----WKTSYGFLAGIIIVVVIVLEILAYL- 1152
Query: 377 VIKRKKSENK 386
K+SE +
Sbjct: 1153----KRSEKR 1170
>TC82242 weakly similar to GP|21592781|gb|AAM64730.1 putative membrane
protein {Arabidopsis thaliana}, partial (9%)
Length = 580
Score = 89.7 bits (221), Expect = 2e-18
Identities = 48/118 (40%), Positives = 67/118 (56%), Gaps = 2/118 (1%)
Frame = +2
Query: 281 THRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEAL 340
THR GI+IF T+Q+ A L+PK R YWN+YH +GY + I +INIFKG L
Sbjct: 14 THRDFGILIFTFSTIQMLAFRLKPKSTDDYRKYWNMYHHFLGYGLLAIIVINIFKGINIL 193
Query: 341 EVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWI--IVIKRKKSENKLQGMNGTNGN 396
+ +W+ +Y GI+ LG +A LE +TWI I+ K KK ++ + N N N
Sbjct: 194 HGGS-----SWRWSYIGILIGLGTIAFALEIFTWIKFIMDKWKKDKHHDKKENQPNKN 352
>TC77701 similar to PIR|T47789|T47789 hypothetical protein F17J16.120 -
Arabidopsis thaliana, partial (4%)
Length = 961
Score = 89.0 bits (219), Expect = 3e-18
Identities = 59/199 (29%), Positives = 96/199 (47%), Gaps = 4/199 (2%)
Frame = +1
Query: 11 ISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRH 70
IS +F++ + C SQ +N +T+C DLP L++ LH++Y+ T + IAF
Sbjct: 58 ISTCIFISLITPSHSALKCASQKLPSNRSYTNCTDLPSLSATLHFSYNTTNHSIAIAF-- 231
Query: 71 KGITDTNR--WVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESN 128
T N+ WV+W INP M GAQAL+A +++G YT ++ + +
Sbjct: 232 -SATPKNKDDWVSWGINPT---GGKMVGAQALIA-YKTNGNVGVYTYNLTSFGGINEVKS 396
Query: 129 ISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQSKE 188
+S GL A N +TI+A + LP + ++ +WQ G + P H N +
Sbjct: 397 LSVETWGLSAEESNGVITIFAGVKLPEKSDNVTQVWQVGPVVAGKPGKHLFEKENLNAFT 576
Query: 189 SLDLRSGASEQG--SGGGS 205
+L + + G S GG+
Sbjct: 577 ALSVVGSTTVGGANSTGGA 633
>TC88786 weakly similar to GP|7268619|emb|CAB78828.1 unknown protein
{Arabidopsis thaliana}, partial (22%)
Length = 1166
Score = 62.8 bits (151), Expect = 2e-10
Identities = 38/131 (29%), Positives = 63/131 (48%), Gaps = 4/131 (3%)
Frame = +1
Query: 213 HGVLNAISWGILMPLGAVIARYLKVFKSADPAW----FYLHVTCQSAAYIVGVAGWGTGL 268
HG L S G LMP+G + R + +P W FY+H Q A ++ AG +
Sbjct: 442 HGFLLWASMGFLMPIGILAIRLSN--REENPRWLRILFYVHTIFQVIAVLLATAGAIMSI 615
Query: 269 KLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIII 328
K + + + H+ LG+ ++ + LQV + RP++ K R W HW +G A +
Sbjct: 616 K---NFNNLFNNNHQRLGVALYGVIWLQVLVGIFRPQRGSKRRSVWFFAHWILGTAVTFL 786
Query: 329 SIINIFKGFEA 339
++N++ G A
Sbjct: 787 GVLNVYIGLAA 819
>BF639962 weakly similar to GP|21593814|gb| unknown {Arabidopsis thaliana},
partial (20%)
Length = 473
Score = 60.5 bits (145), Expect = 1e-09
Identities = 32/93 (34%), Positives = 49/93 (52%), Gaps = 1/93 (1%)
Frame = +3
Query: 11 ISILMFVTTTLAQTTTQ-TCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFR 69
IS L F+ + + T + TC +Q T ++ +C DLP L S+LH+T+D + L + F
Sbjct: 108 ISFLFFLLSMFSATVSSLTCSTQKLTGTKVYPNCIDLPVLNSFLHYTHDTSNSTLSVVFV 287
Query: 70 HKGITDTNRWVAWAINPNNDLASSMNGAQALVA 102
W++W INP A+ M GA +VA
Sbjct: 288 ATP-PSPGGWISWGINPT---ATGMVGAHVIVA 374
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 30.8 bits (68), Expect = 0.97
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = -1
Query: 99 ALVAILQSSGTPKAYTSSIANSRTQLAESNISYP 132
+L I S TPK YTS + S+ +LA S +YP
Sbjct: 1326 SLPVIASRSNTPKLYTSLFSFSKPELAYSGATYP 1225
>TC83498 similar to GP|10120315|emb|CAC08184. beta-ketoacyl-acyl carrier
protein synthase III {Pisum sativum}, partial (44%)
Length = 756
Score = 29.6 bits (65), Expect = 2.2
Identities = 28/81 (34%), Positives = 34/81 (41%), Gaps = 2/81 (2%)
Frame = +2
Query: 280 STHRTLGIVIFCLGTLQ-VFALLLRPKKDHKIRF-YWNLYHWGVGYATIIISIINIFKGF 337
+THR +FC GT++ A L P K K RF G G A + I N
Sbjct: 215 TTHRFQAFKVFCSGTIEGAAASTLPPSKSPKPRFLIQGCKLVGCGSALPSLQITN----- 379
Query: 338 EALEVSAADRYDNWKHAYTGI 358
E L D D W +A TGI
Sbjct: 380 EDLS-KVVDTSDEWIYARTGI 439
>TC76905 similar to PIR|T48939|T48939 acidic ribosomal protein P2-like -
Arabidopsis thaliana, complete
Length = 683
Score = 28.9 bits (63), Expect = 3.7
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Frame = -3
Query: 5 LRFMFLISILMFV--TTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTY 57
+RF FL+ +L+F+ ++ + TTT+ C S + T C+ P ++ L + +
Sbjct: 420 IRFFFLLYLLLFLCLSSCRSSTTTRRCSSNSDTTTTRRN*CQLFPTRSNNLSYIF 256
>AJ502588 similar to GP|7268619|emb| unknown protein {Arabidopsis thaliana},
partial (8%)
Length = 555
Score = 28.9 bits (63), Expect = 3.7
Identities = 23/84 (27%), Positives = 37/84 (43%), Gaps = 6/84 (7%)
Frame = +1
Query: 213 HGVLNAISWGILMPLGAVIARYLKVFKSADPA------WFYLHVTCQSAAYIVGVAGWGT 266
HG+L S G LMPLG + ++ A+P FY HV Q + ++ G
Sbjct: 307 HGLLLWGSVGFLMPLGILT---IRGSNKAEPGSRRSRILFYFHVAFQMLSVLLATVGAAM 477
Query: 267 GLKLGSDSAGVTYSTHRTLGIVIF 290
L +S + H+ LG+ ++
Sbjct: 478 SLMKFENSFD---NNHQRLGLALY 540
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.133 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,734,041
Number of Sequences: 36976
Number of extensions: 175369
Number of successful extensions: 933
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 916
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 924
length of query: 402
length of database: 9,014,727
effective HSP length: 98
effective length of query: 304
effective length of database: 5,391,079
effective search space: 1638888016
effective search space used: 1638888016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148482.6


