

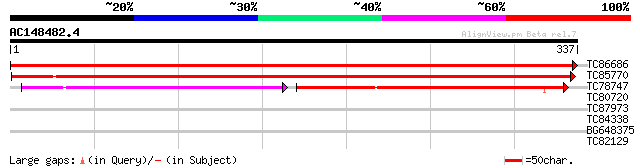
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148482.4 + phase: 0
(337 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86686 similar to GP|16226301|gb|AAL16128.1 AT5g47540/MNJ7_13 {... 657 0.0
TC85770 similar to GP|16226301|gb|AAL16128.1 AT5g47540/MNJ7_13 {... 551 e-157
TC78747 similar to GP|21593944|gb|AAM65898.1 unknown {Arabidopsi... 164 6e-67
TC80720 similar to PIR|E96635|E96635 hypothetical protein T7P1.1... 32 0.35
TC87973 weakly similar to PIR|D86282|D86282 protein F10B6.26 [im... 29 2.9
TC84338 weakly similar to GP|1495247|emb|CAA66220.1 orf 05 {Arab... 28 5.0
BG648375 weakly similar to PIR|T48624|T486 hypothetical protein ... 28 6.5
TC82129 weakly similar to GP|12002439|gb|AAG43351.1 chalcone syn... 27 8.5
>TC86686 similar to GP|16226301|gb|AAL16128.1 AT5g47540/MNJ7_13 {Arabidopsis
thaliana}, partial (96%)
Length = 1527
Score = 657 bits (1696), Expect = 0.0
Identities = 337/337 (100%), Positives = 337/337 (100%)
Frame = +1
Query: 1 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNS 60
MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNS
Sbjct: 112 MKGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNS 291
Query: 61 ESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASD 120
ESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASD
Sbjct: 292 ESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASD 471
Query: 121 YLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNF 180
YLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNF
Sbjct: 472 YLENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNF 651
Query: 181 DIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML 240
DIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML
Sbjct: 652 DIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDML 831
Query: 241 LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVA 300
LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVA
Sbjct: 832 LDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVA 1011
Query: 301 NKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLEA 337
NKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLEA
Sbjct: 1012NKSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLEA 1122
>TC85770 similar to GP|16226301|gb|AAL16128.1 AT5g47540/MNJ7_13 {Arabidopsis
thaliana}, partial (86%)
Length = 1537
Score = 551 bits (1420), Expect = e-157
Identities = 278/335 (82%), Positives = 307/335 (90%)
Frame = +1
Query: 2 KGLFKPKPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSE 61
K LFKPKPRTP+DIVR TRDLL +N+ S D+KR+ ++M EL KN+RE+K+ILYGNSE
Sbjct: 232 KSLFKPKPRTPSDIVRNTRDLLRLL-QNSNSSDNKRDNEKMIELFKNLREMKTILYGNSE 408
Query: 62 SEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDY 121
+EPV EAC+QLTQEFF ENTLRLL+ C+PKLNLEARKDATQVVANLQRQ VQSKLIASDY
Sbjct: 409 AEPVPEACSQLTQEFFNENTLRLLVHCLPKLNLEARKDATQVVANLQRQQVQSKLIASDY 588
Query: 122 LENNMDLMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNSPHMKKFFDYIQLPNFD 181
LE N+DLMDIL+ YEN +MALHYGAMLRECIRHQIVAKYVL SPHMKKFFDYIQLPNFD
Sbjct: 589 LEKNLDLMDILVSSYENPEMALHYGAMLRECIRHQIVAKYVLESPHMKKFFDYIQLPNFD 768
Query: 182 IAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLL 241
IAADAAATFKELMTRHKSTVAEFLS NYEWFF +YNSKLLESSNYITRR AVKLLGDMLL
Sbjct: 769 IAADAAATFKELMTRHKSTVAEFLSNNYEWFFDEYNSKLLESSNYITRRQAVKLLGDMLL 948
Query: 242 DRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPADIVSIFVAN 301
DRSN+AVMT+YVSSR+NLRILMNL+RESSKSIQIEAFHVFKLFAANQ KPA+I+ I VAN
Sbjct: 949 DRSNAAVMTRYVSSRDNLRILMNLMRESSKSIQIEAFHVFKLFAANQKKPAEIIGILVAN 1128
Query: 302 KSKMLRLLDEFKIDKEDEQFEADKAQVMEEIASLE 336
+SK+LRLL + KIDKEDEQFEADKAQVM+EIA+LE
Sbjct: 1129RSKLLRLLGDLKIDKEDEQFEADKAQVMKEIAALE 1233
>TC78747 similar to GP|21593944|gb|AAM65898.1 unknown {Arabidopsis
thaliana}, partial (96%)
Length = 1365
Score = 164 bits (416), Expect(2) = 6e-67
Identities = 81/164 (49%), Positives = 116/164 (70%), Gaps = 2/164 (1%)
Frame = +1
Query: 171 FFDYIQLPNFDIAADAAATFKELMTRHKSTVAEFLSKNYEWFFADYNSKLLESSNYITRR 230
FF Y++LPNFD+A+DA +TFK+L+++H + VAEFL+ +Y+ FF Y KLL S NY+TRR
Sbjct: 658 FFKYVELPNFDVASDAFSTFKDLLSKHATVVAEFLTAHYDEFFDQYE-KLLTSPNYVTRR 834
Query: 231 LAVKLLGDMLLDRSNSAVMTQYVSSRENLRILMNLLRESSKSIQIEAFHVFKLFAANQNK 290
++KLL D LL+ N+ +M +Y+ L+++M LL +SSK+IQ+ AFH+FK+F AN NK
Sbjct: 835 QSIKLLSDFLLESPNAQIMKRYILEVRFLKVMMTLLTDSSKNIQLSAFHIFKIFVANPNK 1014
Query: 291 PADIVSIFVANKSKMLRLLDEFKIDK--EDEQFEADKAQVMEEI 332
P D+ I NK K+L LL K EDEQFE +K +++EI
Sbjct: 1015PRDVKIILGKNKGKLLELLHNLSPGKGSEDEQFEEEKEFIIKEI 1146
Score = 107 bits (267), Expect(2) = 6e-67
Identities = 58/158 (36%), Positives = 94/158 (58%)
Frame = +3
Query: 8 KPRTPTDIVRQTRDLLLFFDRNTESRDSKREEKQMTELCKNIRELKSILYGNSESEPVSE 67
+P+TP ++V+ ++ L+ D T + K EK + E+ KN +++++ G+ ESEP +
Sbjct: 171 RPKTPQEVVKSIKESLMALDTKTVV-EVKALEKALEEVEKNFVTMRTMISGDGESEPNLD 347
Query: 68 ACAQLTQEFFKENTLRLLIKCIPKLNLEARKDATQVVANLQRQPVQSKLIASDYLENNMD 127
+QL +E KE+ L LLI +P L EARKD L +Q V S +Y+ +++
Sbjct: 348 QVSQLVEEICKEDVLTLLIHKLPILGWEARKDLVHCWTILLKQKVDSNDCCVEYIHQHIE 527
Query: 128 LMDILIVGYENTDMALHYGAMLRECIRHQIVAKYVLNS 165
L+D L+ Y+N ++AL G MLRECI+ +AKY+L S
Sbjct: 528 LLDFLVACYDNKEIALSSGIMLRECIKFPNLAKYILES 641
>TC80720 similar to PIR|E96635|E96635 hypothetical protein T7P1.13
[imported] - Arabidopsis thaliana, partial (46%)
Length = 822
Score = 32.0 bits (71), Expect = 0.35
Identities = 17/53 (32%), Positives = 33/53 (62%), Gaps = 4/53 (7%)
Frame = -3
Query: 258 NLRILMNLLRESSKSIQIEAFHVFKLFAANQNKPA----DIVSIFVANKSKML 306
N+++LM+ + S+K ++AFH F + QNK A DI+++++ +K +L
Sbjct: 775 NIKMLMSAIWFSNKIT*VQAFHHLATFGSRQNK*AGLLRDILNLYLISKKYVL 617
>TC87973 weakly similar to PIR|D86282|D86282 protein F10B6.26 [imported] -
Arabidopsis thaliana, partial (82%)
Length = 1073
Score = 28.9 bits (63), Expect = 2.9
Identities = 19/75 (25%), Positives = 31/75 (41%)
Frame = -3
Query: 197 HKSTVAEFLSKNYEWFFADYNSKLLESSNYITRRLAVKLLGDMLLDRSNSAVMTQYVSSR 256
H +T+ KN + F S L S Y + L L + + S+ T ++
Sbjct: 390 HSTTLLSTYQKNLYFLFL--GSILFVSIRYASHNLTTSLFITLQMSTSSRRDQTHHILQL 217
Query: 257 ENLRILMNLLRESSK 271
+I+MN LRE +
Sbjct: 216 LREKIIMNFLREKQR 172
>TC84338 weakly similar to GP|1495247|emb|CAA66220.1 orf 05 {Arabidopsis
thaliana}, partial (9%)
Length = 670
Score = 28.1 bits (61), Expect = 5.0
Identities = 18/65 (27%), Positives = 36/65 (54%), Gaps = 7/65 (10%)
Frame = +1
Query: 31 ESRDSKREEKQMTE----LCKNIRELKSILYGNSES---EPVSEACAQLTQEFFKENTLR 83
E++ SK E +++ + LCKN+R + ++ G+ +S P+ +L++ E TL
Sbjct: 289 EAKSSKVECRELAKKVEILCKNLRGVVRVVTGSHQSLNDRPIRRMVRELSKNL--EKTLA 462
Query: 84 LLIKC 88
L+ +C
Sbjct: 463 LVRRC 477
>BG648375 weakly similar to PIR|T48624|T486 hypothetical protein F18O22.300 -
Arabidopsis thaliana, partial (19%)
Length = 796
Score = 27.7 bits (60), Expect = 6.5
Identities = 13/31 (41%), Positives = 20/31 (63%)
Frame = -3
Query: 155 HQIVAKYVLNSPHMKKFFDYIQLPNFDIAAD 185
H+ + + +L SPH+ K YI L +FDI A+
Sbjct: 779 HRRLTEVILASPHLSKSILYIFLCSFDI*AE 687
>TC82129 weakly similar to GP|12002439|gb|AAG43351.1 chalcone synthase
{Arabidopsis korshinskyi}, partial (30%)
Length = 976
Score = 27.3 bits (59), Expect = 8.5
Identities = 19/71 (26%), Positives = 34/71 (47%)
Frame = +1
Query: 40 KQMTELCKNIRELKSILYGNSESEPVSEACAQLTQEFFKENTLRLLIKCIPKLNLEARKD 99
K++ ++++ LK E+ + E LT+EF K+NT + +P NL
Sbjct: 139 KEVNNDDEHLQRLKLKFKSICENSKIEERHVALTEEFLKQNTEDGKYESLPLENL----- 303
Query: 100 ATQVVANLQRQ 110
T+ V NL ++
Sbjct: 304 PTEQVVNLAKE 336
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,840,541
Number of Sequences: 36976
Number of extensions: 86516
Number of successful extensions: 404
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 401
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 401
length of query: 337
length of database: 9,014,727
effective HSP length: 97
effective length of query: 240
effective length of database: 5,428,055
effective search space: 1302733200
effective search space used: 1302733200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148482.4


