

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148406.6 + phase: 0
(569 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
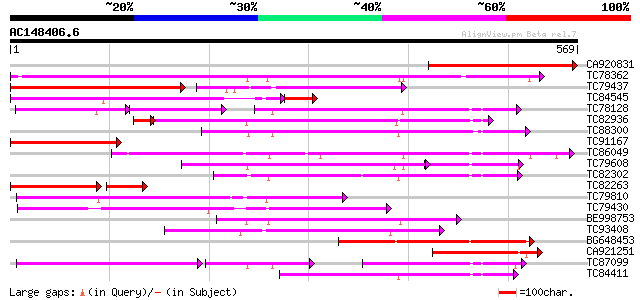
Score E
Sequences producing significant alignments: (bits) Value
CA920831 weakly similar to GP|14209546|dbj putative nitrate tran... 308 3e-84
TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter... 302 3e-82
TC79437 similar to PIR|G96720|G96720 nitrate transporter (NTL1) ... 157 1e-75
TC84545 weakly similar to GP|14209546|dbj|BAB56042. putative nit... 237 2e-72
TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T1... 186 3e-66
TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptid... 220 4e-59
TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affini... 221 6e-58
TC91167 weakly similar to GP|20147231|gb|AAM10330.1 At1g68570/F2... 215 3e-56
TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative tra... 213 1e-55
TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {... 158 2e-54
TC82302 similar to PIR|F84663|F84663 probable nitrate transporte... 208 5e-54
TC82263 weakly similar to SP|Q16348|PET2_HUMAN Oligopeptide tran... 155 1e-52
TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter... 203 2e-52
TC79430 similar to PIR|F86358|F86358 Similar to peptide transpor... 195 3e-50
BE998753 similar to PIR|T45958|T45 oligopeptide transporter-like... 191 6e-49
TC93408 similar to GP|11933407|dbj|BAB19758. putative nitrate tr... 190 1e-48
BG648453 similar to GP|15293215|gb putative oligopeptide transpo... 178 4e-45
CA921251 weakly similar to GP|21740678|emb OSJNBa0084K20.21 {Ory... 175 5e-44
TC87099 similar to PIR|G86449|G86449 hypothetical protein AAF813... 121 2e-42
TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersico... 159 3e-39
>CA920831 weakly similar to GP|14209546|dbj putative nitrate transporter
{Oryza sativa (japonica cultivar-group)}, partial (11%)
Length = 726
Score = 308 bits (790), Expect = 3e-84
Identities = 149/149 (100%), Positives = 149/149 (100%)
Frame = -1
Query: 421 FFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQNN 480
FFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQNN
Sbjct: 723 FFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQNN 544
Query: 481 LNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSKELGETHLLMVGGRKHDDKAKAK 540
LNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSKELGETHLLMVGGRKHDDKAKAK
Sbjct: 543 LNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSKELGETHLLMVGGRKHDDKAKAK 364
Query: 541 ESSQTSEANTEGPSSSDETDDGKEKERHR 569
ESSQTSEANTEGPSSSDETDDGKEKERHR
Sbjct: 363 ESSQTSEANTEGPSSSDETDDGKEKERHR 277
>TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter {Cucumis
sativus}, partial (88%)
Length = 1871
Score = 302 bits (773), Expect = 3e-82
Identities = 183/561 (32%), Positives = 306/561 (53%), Gaps = 26/561 (4%)
Frame = +3
Query: 2 GFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFG 61
GF NM++ Y ++ L S++NTL+NF G + L L+GAFI+D++ RF T +
Sbjct: 150 GFHANMIT---YLTQQLNMPLVSASNTLSNFSGLSSLTPLLGAFIADSFAGRFWTIVFAT 320
Query: 62 SLEVMALVMITVQAALDNLHPKACGKS-SCVEGGIAVMF--YTSLCLYALGMGGVRGSLT 118
+ + L+ IT A + + P C +C E + ++ Y +L L +LG GG+R +
Sbjct: 321 LIYELGLITITTSAIVPHFRPPPCPTQVNCQEAKSSQLWILYLALFLTSLGSGGIRSCVV 500
Query: 119 AFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVAS 178
F +QFD FNW S+ +T +V++ W WG I T+A
Sbjct: 501 PFSGDQFDMTKKGVESRKWNLFNWYFFCMGFASLSALTIVVYIQDHMGWGWGLGIPTIAM 680
Query: 179 SIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYE-VYKDATIE 237
I + +G Y+ P SP LR+ QVIV AF+ R LP + LY+ + D++I
Sbjct: 681 FIAIIAFLLGSRLYKTLKPSGSPFLRLAQVIVAAFRKRNDALPNDPKLLYQNLELDSSIS 860
Query: 238 ---KIVHTNQMRFLDKAAILQEN-----SESQQPWKVCTVTQVEEVKILTRMLPILASTI 289
++ HT+Q ++LDKAAI+ + ++ W + TV +VEE+K L RMLPI AS I
Sbjct: 861 LEGRLSHTDQYKWLDKAAIVTDEEAKNLNKLPNLWNLATVHRVEELKCLVRMLPIWASGI 1040
Query: 290 VMNTCLAQLQTFSVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRK 348
++ T + +F + Q +M+ L +F + +S+ + ++ + T + +YE FVPFIR+
Sbjct: 1041LLITASSSQHSFVIVQARTMDRHLSHTFEISPASMAIFSVLTMMTGVILYERLFVPFIRR 1220
Query: 349 ITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRK----DPSK---PISLFWLS 401
T +P+G+T LQR+G+G V++ I+ ++ +E+KR+ K D K PIS+FWL
Sbjct: 1221FTKNPAGITCLQRMGIGFVINIIATIVSALLEIKRKKVASKYHLLDSPKAIIPISVFWLV 1400
Query: 402 FQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTK 461
QY + G+A++F VG LEF Y ++P +M+S +T+ +++++G+F+ T+ V T+
Sbjct: 1401PQYFLHGVAEVFMNVGHLEFLYDQSPESMRSSATALYCIAIAIGHFIGTLLV----TLVH 1568
Query: 462 RITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYK--SEEDSNNSKE 519
+ T ++ WL +LN+ L +Y+ + + +NF ++ A Y YK E + NN +
Sbjct: 1569KYTGKERNWLPDRNLNRGRLEYYYFLVCGIQVINFIYYVICAWIYNYKPLEEINENNQGD 1748
Query: 520 L----GETHLLMVGGRKHDDK 536
L GE L+ + K D+K
Sbjct: 1749LEQTNGELSLVSLNEGKVDEK 1811
>TC79437 similar to PIR|G96720|G96720 nitrate transporter (NTL1)
53025-56402 [imported] - Arabidopsis thaliana, partial
(61%)
Length = 1424
Score = 157 bits (398), Expect(2) = 1e-75
Identities = 77/179 (43%), Positives = 113/179 (63%), Gaps = 3/179 (1%)
Frame = +3
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
+ ++ N +LVLY MH S SAN +TNFMG++FLL+L+G F+SD +L + L+
Sbjct: 165 LAYLANASNLVLYLKEYMHMSPSKSANNVTNFMGTSFLLALLGGFLSDAFLTTYHVYLIS 344
Query: 61 GSLEVMALVMITVQAALDNLHPKACGKSS-C--VEGGIAVMFYTSLCLYALGMGGVRGSL 117
+E++ L+++T+QA + +L P C +S+ C V GG A M + L L ALG+GG++GSL
Sbjct: 345 AVIELIGLIILTIQAHVPSLKPTKCNESTPCEEVNGGKAAMLFAGLYLVALGVGGIKGSL 524
Query: 118 TAFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITV 176
G QFDE PN K +TFFN+ + + G++I VT +VWV K W WGF I T+
Sbjct: 525 PVHGGEQFDEATPNGRKQRSTFFNYFVFCLSCGALIAVTFVVWVEDNKGWEWGFAISTI 701
Score = 144 bits (363), Expect(2) = 1e-75
Identities = 84/231 (36%), Positives = 130/231 (55%), Gaps = 20/231 (8%)
Frame = +1
Query: 188 GKPFYRIKTPGESPILRIIQVIVVAFKNR------------KLQLPESN-------EQLY 228
G YR K P SP+ I++V++ A N + SN ++L+
Sbjct: 736 GSTTYRNKVPSGSPLTTILKVLIAATLNSCCTNKNSCSAVVNMSSSPSNPSTQAKEQELH 915
Query: 229 EVYKDATIEKIVHTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILAST 288
+ K T +I + ++FL+KA + S Q CT+ Q+E+VK++ ++ PI A T
Sbjct: 916 QTLKPTTPSQIP-SESLKFLNKAITNKPIHSSLQ----CTIQQLEDVKLVFKIFPIFACT 1080
Query: 289 IVMNTCLAQLQTFSVQQGNSMNLKL-GSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIR 347
I++N CLAQL TFSV+Q +MN L SF VP +S+PV P++FL L PIY+ +P+ R
Sbjct: 1081IMLNACLAQLSTFSVEQAATMNTTLFSSFKVPPASLPVFPVLFLMILAPIYDHIIIPYAR 1260
Query: 348 KITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLF 398
K+T +G+T LQR+G+GL+LS ++M IA +E+K + K I+ F
Sbjct: 1261KVTKSEAGITHLQRIGIGLILSIVAMAIAAIVEIKDKK*HHTQMKKSITYF 1413
>TC84545 weakly similar to GP|14209546|dbj|BAB56042. putative nitrate
transporter {Oryza sativa (japonica cultivar-group)},
partial (15%)
Length = 989
Score = 237 bits (605), Expect(2) = 2e-72
Identities = 125/284 (44%), Positives = 168/284 (59%), Gaps = 8/284 (2%)
Frame = +2
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
M FV N VSL+ YF G M+F L+ SA TLTN MG+ FLL L+G ISDTYL+RF TC++F
Sbjct: 131 MAFVANAVSLITYFTGSMNFSLTKSATTLTNLMGTAFLLPLLGGLISDTYLSRFKTCVLF 310
Query: 61 GSLEVMALVMITVQAALDNLHPKACGKSSCVE--------GGIAVMFYTSLCLYALGMGG 112
S+E++ ++T+QA L P C + ++ G A + YT L L+ALG G
Sbjct: 311 ASMELLGYGILTIQARFQQLRPIPCKDIAPIDMNQCEPANGSQAAILYTGLYLFALGTSG 490
Query: 113 VRGSLTAFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFF 172
V+ +L A GA+QFD+KDP EA L++FFNW LLS T G++ GVT I W+S+ + W+W F
Sbjct: 491 VKAALPALGADQFDDKDPKEAAQLSSFFNWFLLSLTTGAIFGVTFINWISSNQGWYWSFT 670
Query: 173 IITVASSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYK 232
+ T+A + +++ +GK FYR TP SP++RIIQV V AFKN
Sbjct: 671 VCTIAVFLSILSICMGKSFYRNNTPKGSPLIRIIQVFVAAFKN----------------- 799
Query: 233 DATIEKIVHTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVK 276
RFLD+AA+ S W +CTVTQVEE K
Sbjct: 800 -------------RFLDRAAV---GGNSTGSWNLCTVTQVEETK 883
Score = 54.3 bits (129), Expect(2) = 2e-72
Identities = 25/34 (73%), Positives = 29/34 (84%)
Frame = +1
Query: 276 KILTRMLPILASTIVMNTCLAQLQTFSVQQGNSM 309
KIL RMLPI+ STI MNTCLAQLQTFS+QQ ++
Sbjct: 880 KILVRMLPIILSTIFMNTCLAQLQTFSIQQSTTL 981
>TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T17F3_10
{Arabidopsis thaliana}, partial (77%)
Length = 2022
Score = 186 bits (471), Expect(3) = 3e-66
Identities = 107/278 (38%), Positives = 160/278 (57%), Gaps = 10/278 (3%)
Frame = +2
Query: 246 RFLDKAAILQENSESQQ-----PWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQT 300
R LDKAA++ E + W + ++ QVEEVK L R LPI A+ I+ T +AQ T
Sbjct: 1058 RVLDKAALVMEGDLNPDGTIVNQWNLVSIQQVEEVKCLARTLPIWAAGILGFTAMAQQGT 1237
Query: 301 FSVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQL 359
F V Q M+ LG F +PA S+ VI LI + +P Y+ VP +RKIT + G+T L
Sbjct: 1238 FIVSQAMKMDRHLGPKFQIPAGSLGVISLIVIGLWVPFYDRICVPSLRKITKNEGGITLL 1417
Query: 360 QRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPS----KPISLFWLSFQYAIFGIADMFTL 415
QR+G+G+V S ISM +AG +E RRD +P+ P+S+ WL Q + G + F +
Sbjct: 1418 QRIGIGMVFSIISMIVAGLVEKVRRDVANSNPTPQGIAPMSVMWLFPQLVLMGFCEAFNI 1597
Query: 416 VGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFD 475
+GL+EFF R+ P M+S++ + S +L ++S++ V +++VTK T + WL +
Sbjct: 1598 IGLIEFFNRQFPDHMRSIANALFSCSFALANYVSSILVITVHSVTK--THNHPDWLTN-N 1768
Query: 476 LNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEED 513
+N+ L+ FY+ LA + LN FLY + Y YK D
Sbjct: 1769 INEGRLDYFYYLLAGVGVLNLVYFLYVSQRYHYKGSVD 1882
Score = 55.5 bits (132), Expect(3) = 3e-66
Identities = 32/97 (32%), Positives = 45/97 (45%)
Frame = +3
Query: 121 GANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSI 180
G +QFD K + +FFNW S T+ + T IV++ +W +GF I T+
Sbjct: 666 GVDQFDPTTEKGKKGINSFFNWYYTSFTVVLLFTQTVIVYIQDSISWKFGFAIPTLCMLX 845
Query: 181 GFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRK 217
+ IG Y P S I QV+V +FK RK
Sbjct: 846 SIILFFIGTKIYVHVKPEGSIFSSIAQVLVASFKKRK 956
Score = 50.4 bits (119), Expect(3) = 3e-66
Identities = 31/122 (25%), Positives = 54/122 (43%), Gaps = 7/122 (5%)
Frame = +2
Query: 7 MVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVM 66
+ + ++Y H + ++N L + G + L+GAFISDTY RF T ++
Sbjct: 302 LANFMVYLTREFHLEQVHASNILNIWGGISNFAPLLGAFISDTYTGRFKTIAFASFFSLL 481
Query: 67 ALVMITVQAALDNLHPKAC-----GKSSCVEGGIAVM--FYTSLCLYALGMGGVRGSLTA 119
+ +T+ A L L P +C + CV + + + L ++G G+R
Sbjct: 482 GMTAVTLTAWLPKLQPPSCTPQQQALNQCVTANSSQVGFLFMGLIFLSIGSSGIRPCSIP 661
Query: 120 FG 121
FG
Sbjct: 662 FG 667
>TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptide
transporter [imported] - Arabidopsis thaliana, partial
(53%)
Length = 1112
Score = 220 bits (561), Expect(2) = 4e-59
Identities = 130/354 (36%), Positives = 203/354 (56%), Gaps = 13/354 (3%)
Frame = +3
Query: 145 LSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFVTLAIGKPFYRIKT-PGESPIL 203
L S LG++I G+V++ W G+ I T + V IG P YR K +SP
Sbjct: 63 LXSFLGALIATLGLVYIQENLGWGLGYGIPTAGLILSLVIFYIGTPIYRHKVRTSKSPAK 242
Query: 204 RIIQVIVVAFKNRKLQLPESNEQLYEVYKDATI----EKIVHTNQMRFLDKAAILQENSE 259
II+V +VAFK+RKLQLP + +L+E + + ++ HT +RFLDKAAI ++ +
Sbjct: 243 DIIRVFIVAFKSRKLQLPSNPSELHEFQMEHCVIRGKRQVYHTPTLRFLDKAAIKEDPTT 422
Query: 260 SQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKLG-SFTV 318
TV QVE K++ ML I T++ +T AQ+ T V+QG +++ LG F +
Sbjct: 423 GSSRRVPMTVNQVEGAKLILGMLLIWLVTLIPSTIWAQINTLFVKQGTTLDRNLGPDFKI 602
Query: 319 PASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGF 378
PA+S+ + + +P+Y+ FVPF+R+ T HP G+T LQR+G+G + I++ IA
Sbjct: 603 PAASLGSFVTLSMLLSVPMYDRLFVPFMRQKTGHPRGITLLQRLGIGFSIQIIAIAIAYA 782
Query: 379 IEVKRRDQ-------GRKDPSKPISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMK 431
+EV+R G KD P+S+FWL QY + GIAD+F +GLLEFFY ++P M+
Sbjct: 783 VEVRRMHVIKENHIFGPKD-IVPMSIFWLLPQYVLIGIADVFNAIGLLEFFYDQSPEDMQ 959
Query: 432 SLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQNNLNLFY 485
SL T+F + +G FL++ V + + +T R ++ W+ +LN ++L +Y
Sbjct: 960 SLGTTFFTSGIGVGNFLNSFLVTMTDKITGR--GDRKSWIAD-NLNDSHLXYYY 1112
Score = 26.6 bits (57), Expect(2) = 4e-59
Identities = 9/22 (40%), Positives = 16/22 (71%)
Frame = +2
Query: 125 FDEKDPNEAKALATFFNWLLLS 146
FD+ +P+E + A+FFNW + +
Sbjct: 2 FDDFNPHEKELKASFFNWWMFT 67
>TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affinity nitrate
transporter {Nicotiana plumbaginifolia}, partial (56%)
Length = 1169
Score = 221 bits (563), Expect = 6e-58
Identities = 134/349 (38%), Positives = 196/349 (55%), Gaps = 19/349 (5%)
Frame = +1
Query: 193 RIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEV--YKDATIEK----IVHTNQMR 246
R K SP+ +I V V A++ R ++LP + L+ V +D + K + H+ Q R
Sbjct: 40 RFKKLAGSPLTQIAVVYVAAWRKRNMELPYDSSLLFNVDDIEDEMLRKKKQVLPHSKQFR 219
Query: 247 FLDKAAILQENSESQQ-----PWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTF 301
FLDKAAI ++ + W + T+T VEEVK++ RMLPI A+TI+ T AQ+ TF
Sbjct: 220 FLDKAAIKDPKTDGNEINVVRKWYLSTLTDVEEVKLVLRMLPIWATTIMFWTVYAQMTTF 399
Query: 302 SVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQ 360
SV Q ++N +G SF +P +S+ + + IPIY+ VP RKI +P G+T LQ
Sbjct: 400 SVSQATTLNRHIGKSFQIPPASLTAFFIGSILLTIPIYDRVIVPITRKIFKNPQGLTPLQ 579
Query: 361 RVGVGLVLSAISMTIAGFIEVKRRDQGR-----KDPSK--PISLFWLSFQYAIFGIADMF 413
R+GVGLV S +M A E+KR +P+ P+S+FWL Q+ G + F
Sbjct: 580 RIGVGLVFSIFAMVAAALTELKRMRMAHLHNLTHNPNSEIPMSVFWLIPQFFFVGSGEAF 759
Query: 414 TLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQG 473
T +G L+FF RE P MK++ST ++SLG+F S++ V +++ VT + P WL
Sbjct: 760 TYIGQLDFFLRECPKGMKTMSTGLFLSTLSLGFFFSSLLVTLVHKVTSQHKP----WLAD 927
Query: 474 FDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSKELGE 522
+LN+ L FYW LA+LS LN +L A+WY YK + + EL E
Sbjct: 928 -NLNEAKLYNFYWLLALLSVLNLVIYLLCANWYVYKDKRLAEEGIELEE 1071
>TC91167 weakly similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7
{Arabidopsis thaliana}, partial (5%)
Length = 728
Score = 215 bits (548), Expect = 3e-56
Identities = 109/112 (97%), Positives = 109/112 (97%)
Frame = +2
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF
Sbjct: 356 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 535
Query: 61 GSLEVMALVMITVQAALDNLHPKACGKSSCVEGGIAVMFYTSLCLYALGMGG 112
GSLEVMALVMITVQAALDNLHPKACGKSSCVEGGIAVMFYTSL LYA G GG
Sbjct: 536 GSLEVMALVMITVQAALDNLHPKACGKSSCVEGGIAVMFYTSLXLYAFGNGG 691
>TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative transport
protein {Arabidopsis thaliana}, partial (43%)
Length = 1836
Score = 213 bits (543), Expect = 1e-55
Identities = 146/494 (29%), Positives = 254/494 (50%), Gaps = 30/494 (6%)
Frame = +2
Query: 103 LCLYALGMGGVRGSLTAFGANQFDEKD-PNEAKALATFFNWLLLSSTLGSVIGVTGIVWV 161
L L A+G GG+ SL+ FGA Q + KD P+ + L FF+W +T+ +I +TGIV++
Sbjct: 2 LILIAIGNGGITCSLS-FGAYQVNRKDNPDGYRVLEIFFSWYYAFTTIAVIIALTGIVYI 178
Query: 162 STQKAWHWGFFIITVASSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLP 221
W GF + + I V + P Y S QV V A+KNRKL LP
Sbjct: 179 QDHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLP 358
Query: 222 -ESNEQLYEVYKDATIEKIVHTNQMRFLDKAAILQENSE-------SQQPWKVCTVTQVE 273
+++ + Y KD+ E +V T+++RFL+KA +++++ + + W +CTV QVE
Sbjct: 359 PKTSPEFYHQQKDS--ELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVE 532
Query: 274 EVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMN---LKLGSFTVPASSIPVIPLIF 330
E+K + +++P+ ++ I M+ + +F + Q S++ + +F VPA S VI ++
Sbjct: 533 ELKAIIKVIPLWSTAITMSINIGG--SFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVA 706
Query: 331 LCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKD 390
+ I IY+ +P KI P ++ +R+G+GL + + + A E RR + K+
Sbjct: 707 ILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKE 886
Query: 391 PSK-------PISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMS 443
+S WL+ Q + GIA+MF ++G EF+Y+E P +M S++ S + L+M
Sbjct: 887 GYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMG 1066
Query: 444 LGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWA 503
+G +S++ +++I + T + +GW+ ++N+ + + +YW + ++ LN +L +
Sbjct: 1067VGNLVSSLVLSIIESTTP--SGGNEGWVSD-NINKGHFDKYYWVIVGINALNLLYYLVCS 1237
Query: 504 SWYKYKSEEDSNNSKELG----ETHLLMVGGRKHDDKAKAKESSQTSE-------ANTEG 552
Y +E SN SKE G E+ DDK + SS+ E A E
Sbjct: 1238WAYGPTVDEVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEK 1417
Query: 553 PSSSDETDDGKEKE 566
+ E D KE++
Sbjct: 1418VFKNSEQRDLKEED 1459
>TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {Arabidopsis
thaliana}, partial (59%)
Length = 1489
Score = 158 bits (399), Expect(2) = 2e-54
Identities = 97/264 (36%), Positives = 155/264 (57%), Gaps = 14/264 (5%)
Frame = +2
Query: 173 IITVASSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYK 232
I T+A I + G P YR P SP R+++V V AF+ +K+ ++ LY+ +
Sbjct: 8 IPTLAMLISIIAFIGGYPLYRNLNPEGSPFTRLVKVGVAAFRKKKIPKVPNSTLLYQNDE 187
Query: 233 -DATIE---KIVHTNQMRFLDKAAILQENSESQQP--WKVCTVTQVEEVKILTRMLPILA 286
DA+I K++H++Q++FLDKAAI+ E ++ P W++ TV +VEE+K + RM PI A
Sbjct: 188 LDASITLGGKLLHSDQLKFLDKAAIVTEEDNTKTPDLWRLSTVHRVEELKSIIRMGPIWA 367
Query: 287 STIVMNTCLAQLQTFSVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPF 345
S I++ T AQ TFS+QQ +MN L SF +PA S+ V ++ + +Y+ +
Sbjct: 368 SGILLITAYAQQGTFSLQQAKTMNRHLTKSFEIPAGSMSVFTILTMLFTTALYDRVLIRV 547
Query: 346 IRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRR----DQGRKDPSK---PISLF 398
R+ T G+T L R+G+G V+S + +AGF+E+KR+ + G + S PIS+F
Sbjct: 548 ARRFTGLDRGITFLHRMGIGFVISLFATFVAGFVEMKRKKVAMEHGLIEHSSEIIPISVF 727
Query: 399 WLSFQYAIFGIADMFTLVGLLEFF 422
WL QY++ G+A+ F +G + F
Sbjct: 728 WLVPQYSLHGMAEAFMSIGAFKSF 799
Score = 73.2 bits (178), Expect(2) = 2e-54
Identities = 39/109 (35%), Positives = 60/109 (54%), Gaps = 10/109 (9%)
Frame = +1
Query: 417 GLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDL 476
G+ EFFY +AP +M S + +F + S+SLG ++ST+ V +++ TK P+ WL +L
Sbjct: 784 GI*EFFYDQAPESMTSTAMAFFWTSISLGNYISTLLVTLVHKFTK--GPNGTNWLPDNNL 957
Query: 477 NQNNLNLFYWFLAILSCLNFFNFLYWASWYKYK----------SEEDSN 515
N+ L FYW + +L +N +L A Y YK S ED+N
Sbjct: 958 NKGKLEYFYWLITLLQFINLIYYLICAKMYTYKQIQVHHKGENSSEDNN 1104
>TC82302 similar to PIR|F84663|F84663 probable nitrate transporter
[imported] - Arabidopsis thaliana, partial (51%)
Length = 1133
Score = 208 bits (529), Expect = 5e-54
Identities = 126/322 (39%), Positives = 187/322 (57%), Gaps = 12/322 (3%)
Frame = +3
Query: 205 IIQVIVVAFKNRKLQLPESNEQLYE-VYKDATIEKIVHTNQMRFLDKAAILQENS----- 258
I QVIV + K RK++LP + LYE +D+ IE+ + Q RFL+KAAI+ E
Sbjct: 3 IFQVIVASIKKRKMELPYNVGSLYEDTPEDSRIEQ---SEQFRFLEKAAIVVEGDFDKDL 173
Query: 259 --ESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKLGSF 316
PWK+C++T+VEEVK++ R+LPI A+TI+ T AQ+ TFSV+Q +M +G+F
Sbjct: 174 YGSGPNPWKLCSLTRVEEVKMMVRLLPIWATTIIFWTTYAQMITFSVEQAATMERNVGNF 353
Query: 317 TVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIA 376
+PA S+ V ++ + + + + +P +K+ P G + LQR +GL+LS I M A
Sbjct: 354 QIPAGSLTVFFVVAILLTLAVNDRIIMPLWKKLNGKP-GFSNLQRNAIGLLLSTIGMAAA 530
Query: 377 GFIEVKRRDQGR----KDPSKPISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKS 432
IEVKR + + PIS+F L Q+ + G + F G L+FF ++P MK+
Sbjct: 531 SLIEVKRLSVAKGVKGNQTTLPISVFLLVPQFFLVGSGEAFIYTGQLDFFITQSPKGMKT 710
Query: 433 LSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILS 492
+ST ++SLG+F+S+ V+V+ VT T QGWL +N+ L+LFY L ILS
Sbjct: 711 MSTGLFLTTLSLGFFVSSFLVSVVKKVTG--TRDGQGWLAD-HINKGRLDLFYALLTILS 881
Query: 493 CLNFFNFLYWASWYKYKSEEDS 514
+NF FL A WYK K + S
Sbjct: 882 FINFVAFLVCAFWYKPKKPKPS 947
>TC82263 weakly similar to SP|Q16348|PET2_HUMAN Oligopeptide transporter
kidney isoform (Peptide transporter 2) (Kidney
H+/peptide cotransporter), partial (3%)
Length = 950
Score = 155 bits (393), Expect(2) = 1e-52
Identities = 76/92 (82%), Positives = 82/92 (88%)
Frame = +1
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
MGFV NMVSLVLYF GVMHFD+ SSANTLTNFMGSTFLLSLVG FISDTYLNRFTTCL+F
Sbjct: 535 MGFVANMVSLVLYFYGVMHFDIPSSANTLTNFMGSTFLLSLVGGFISDTYLNRFTTCLLF 714
Query: 61 GSLEVMALVMITVQAALDNLHPKACGKSSCVE 92
GSLEV+AL ++T QAA D+LHP ACGKSS E
Sbjct: 715 GSLEVLALALVTFQAASDHLHPNACGKSSLCE 810
Score = 69.3 bits (168), Expect(2) = 1e-52
Identities = 33/41 (80%), Positives = 38/41 (92%)
Frame = +3
Query: 98 MFYTSLCLYALGMGGVRGSLTAFGANQFDEKDPNEAKALAT 138
MFYTSL L ALG+GGVRGS+TAFGA+QF+EKD NEAKALA+
Sbjct: 828 MFYTSLSLLALGIGGVRGSMTAFGADQFEEKDSNEAKALAS 950
>TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter
{Arabidopsis thaliana}, partial (60%)
Length = 1236
Score = 203 bits (516), Expect = 2e-52
Identities = 115/343 (33%), Positives = 190/343 (54%), Gaps = 11/343 (3%)
Frame = +3
Query: 8 VSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMA 67
++LV Y +G +H +SSA +TNFMG+ LL L+G F++D L R+ T + ++ +
Sbjct: 216 MNLVTYLVGDLHLHSASSATIVTNFMGTLNLLGLLGGFLADAKLGRYLTVAISATIATVG 395
Query: 68 LVMITVQAALDNLHPKACGK-----SSCVE--GGIAVMFYTSLCLYALGMGGVRGSLTAF 120
+ M+T+ + ++ P C + C++ G + + +L ALG GG++ +++ F
Sbjct: 396 VCMLTLATTIPSMTPPPCSEVRRQHHQCIQASGKQLSLLFAALYTIALGGGGIKSNVSGF 575
Query: 121 GANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSI 180
G++QFD DP E K + FFN ++GS+ V +V+V WG+ I +
Sbjct: 576 GSDQFDTNDPKEEKNMIFFFNRFYFFISIGSLFSVVVLVYVQDNIGRGWGYGISAGTMLV 755
Query: 181 GFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIEKIV 240
L G P YR K P SP+ I +V+ +A+K R L +P S+ L Y +A K+
Sbjct: 756 AVGILLCGTPLYRFKKPQGSPLTIIWRVLFLAWKKRTLPIP-SDPTLLNGYLEA---KVT 923
Query: 241 HTNQMRFLDKAAILQE----NSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLA 296
+T++ R LDKAAIL E + ++ PW V T+TQVEEVK++ ++LPI ++ I+ T +
Sbjct: 924 YTDKFRSLDKAAILDETKSKDGNNENPWLVSTMTQVEEVKMVIKLLPIWSTCILFWTVYS 1103
Query: 297 QLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIFLCTLIPIYE 339
Q+ TF+++Q MN K+GS +PA S+ I + + E
Sbjct: 1104QMNTFTIEQATFMNRKVGSLEIPAGSLSAFLFITILLFTSLNE 1232
>TC79430 similar to PIR|F86358|F86358 Similar to peptide transporter
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1303
Score = 195 bits (496), Expect = 3e-50
Identities = 121/382 (31%), Positives = 203/382 (52%), Gaps = 7/382 (1%)
Frame = +2
Query: 9 SLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMAL 68
+L+ Y G + +++A + + G+ LL L+GAF++D++L R+ T ++ + ++AL
Sbjct: 209 NLINYLTGPLGQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYILAL 388
Query: 69 VMITVQAALDNLHPKACGKSSCVEGGI-AVMFYTSLCLYALGMGGVRGSLTAFGANQFDE 127
++T+ A L + +G A++F+ SL L A GG + + AFGA+QFD
Sbjct: 389 SLLTLSATLPS------------DGDAQAILFFFSLYLVAFAQGGHKPCVQAFGADQFDI 532
Query: 128 KDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFVTLAI 187
P E ++ ++FFNW + T G ++ V+ + +V W GF I +A I ++
Sbjct: 533 NHPQERRSRSSFFNWWYFTFTAGLLVTVSILNYVQDNVGWVLGFGIPWIAMIIALSLFSL 712
Query: 188 GKPFYRIKTPG---ESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIEKIVH--T 242
G YR PG P RI +V + A +N + E ++H +
Sbjct: 713 GTWTYRFSIPGNQQRGPFSRIGRVFITALRNFRTTEEEEPR-----------PSLLHQPS 859
Query: 243 NQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTFS 302
Q FL+KA I + S++ KVC+V++VEE K + R++PI A++++ +Q TF
Sbjct: 860 QQFSFLNKALIASDG--SKENGKVCSVSEVEEAKAILRLVPIWATSLIFAIVFSQSSTFF 1033
Query: 303 VQQGNSMNLK-LGSFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQR 361
+QG +++ K L F VP +S+ + + IP+Y+ VP R T PSG+T LQR
Sbjct: 1034TKQGVTLDRKILPGFYVPPASLQSFISLSIVLFIPVYDRIIVPIARTFTGKPSGITMLQR 1213
Query: 362 VGVGLVLSAISMTIAGFIEVKR 383
+G G++ S ISM IA F+E+KR
Sbjct: 1214IGAGILFSVISMVIAAFVEMKR 1279
>BE998753 similar to PIR|T45958|T45 oligopeptide transporter-like protein -
Arabidopsis thaliana, partial (44%)
Length = 809
Score = 191 bits (485), Expect = 6e-49
Identities = 101/260 (38%), Positives = 157/260 (59%), Gaps = 14/260 (5%)
Frame = +3
Query: 208 VIVVAFKNRKLQLPESNEQLYEVYKDATI----EKIVHTNQMRFLDKAAILQENS---ES 260
VIV + + ++ P LYE+ + K+ HTN++RF DKAA+ E+ ES
Sbjct: 30 VIVASIRKYRVDAPTDKSLLYEIADTESAIKGSRKLDHTNELRFFDKAAVQGESDNLKES 209
Query: 261 QQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKLG--SFTV 318
PW++CTVTQVEE+K + R+LP+ A+ I+ T Q+ T V QG +MN +G SF +
Sbjct: 210 INPWRLCTVTQVEELKSILRLLPVWATGIIFATVYGQMSTLFVLQGQTMNTHVGNSSFKI 389
Query: 319 PASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGF 378
P +S+ + I + +P+Y+ VP RK T H +G+TQLQR+GVGL +S SM A F
Sbjct: 390 PPASLSIFDTISVIFWVPVYDRIIVPIARKFTGHKNGLTQLQRMGVGLFISIFSMVAAAF 569
Query: 379 IEVKRRDQGRKD-----PSKPISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSL 433
+E+ R R++ P+++FW QY + G A++FT +G LEFFY +AP M+SL
Sbjct: 570 LELVRLRTVRRNNYYELEEIPMTIFWQVPQYFLIGCAEVFTFIGQLEFFYEQAPDAMRSL 749
Query: 434 STSFTFLSMSLGYFLSTVFV 453
++ + L+++ G +LS++ V
Sbjct: 750 CSALSLLTVAFGQYLSSLLV 809
>TC93408 similar to GP|11933407|dbj|BAB19758. putative nitrate transporter
NRT1-3 {Glycine max}, partial (44%)
Length = 909
Score = 190 bits (483), Expect = 1e-48
Identities = 105/293 (35%), Positives = 165/293 (55%), Gaps = 12/293 (4%)
Frame = +1
Query: 156 TGIVWVSTQKAWHWGFFIITVASSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKN 215
T +V++ W G+ + T+ +I + G PFYR K P S R+ +VIV + +
Sbjct: 7 TVLVYIQDNVGWTLGYALPTLGLAISIMIFLGGTPFYRHKLPAGSTFTRMARVIVASLRK 186
Query: 216 RKLQLPESNEQLYEV------YKDATIEKIVHTNQMRFLDKAAILQENSESQQPWKVCTV 269
K+ +P+ ++LYE+ K + +I T +RFLDKA++ + S PW +CTV
Sbjct: 187 WKVPVPDDTKKLYELDMEEYAKKGSNTYRIDSTPTLRFLDKASV---KTGSTSPWMLCTV 357
Query: 270 TQVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLI 329
T VEE K + RM+PIL +T V +T +AQ+ T V+QG +++ +GSF +P +S+ +
Sbjct: 358 THVEETKQMLRMIPILVATFVPSTMMAQVNTLFVKQGTTLDRHIGSFKIPPASLAAFVTL 537
Query: 330 FLCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIE------VKR 383
L + +Y+ FFV ++++T +P G+T LQR+G+GLVL I M +A E K
Sbjct: 538 SLLVCVVLYDRFFVRIMQRLTKNPRGITLLQRMGIGLVLHTIIMVVASVTENYRLRVAKE 717
Query: 384 RDQGRKDPSKPISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTS 436
P+S+F L Q+ + G AD F V +EFFY +AP +MKS+ TS
Sbjct: 718 HGLVESGGQVPLSIFILLPQFILMGTADAFLEVAKIEFFYDQAPXSMKSIGTS 876
>BG648453 similar to GP|15293215|gb putative oligopeptide transporter protein
{Arabidopsis thaliana}, partial (29%)
Length = 701
Score = 178 bits (452), Expect = 4e-45
Identities = 83/196 (42%), Positives = 133/196 (67%)
Frame = +1
Query: 331 LCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKD 390
L ++P+Y+ FVPF RKIT H SG++ LQR+G+GL L+ SM A +E KRRD +
Sbjct: 1 LIVVVPLYDTLFVPFARKITGHESGISPLQRIGIGLFLATFSMVSAAVMEKKRRDAA-VN 177
Query: 391 PSKPISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLST 450
++ +S+FW++ Q+ IFG+++MFT VGL+EFFY+++ M++ T+ T+ S S G++LS+
Sbjct: 178 LNQTLSIFWITPQFIIFGLSEMFTAVGLIEFFYKQSLKGMQTFFTAITYCSYSFGFYLSS 357
Query: 451 VFVNVINTVTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKS 510
+ V+++N +T + GWL +LN++ L+LFYW LA+LS LNF N+L+W+ WY Y
Sbjct: 358 LLVSLVNKITS-TSSGGGGWLHDNNLNKDKLDLFYWLLAVLSFLNFINYLFWSRWYSYNP 534
Query: 511 EEDSNNSKELGETHLL 526
+ + + GE H +
Sbjct: 535 SLSTISQE--GEVHAM 576
>CA921251 weakly similar to GP|21740678|emb OSJNBa0084K20.21 {Oryza sativa},
partial (9%)
Length = 751
Score = 175 bits (443), Expect = 5e-44
Identities = 88/114 (77%), Positives = 98/114 (85%), Gaps = 4/114 (3%)
Frame = -1
Query: 425 EAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQNNLNLF 484
E+PS+MKSLSTSFT+LSMS+GYFLSTVFVN+IN VTKRITPSKQGWL GFDLNQ+NLNLF
Sbjct: 745 ESPSSMKSLSTSFTWLSMSIGYFLSTVFVNLINVVTKRITPSKQGWLHGFDLNQSNLNLF 566
Query: 485 YWFLAILSCLNFFNFLYWASWYKYKSEEDSNNS----KELGETHLLMVGGRKHD 534
YWFLAILSCLNFFN+LYWAS YKYKS ED N+S K L E L M+GG K +
Sbjct: 565 YWFLAILSCLNFFNYLYWASRYKYKS-EDQNSSPIGLKSLHEMPLKMIGGTKQN 407
>TC87099 similar to PIR|G86449|G86449 hypothetical protein AAF81343.1
[imported] - Arabidopsis thaliana, partial (86%)
Length = 2205
Score = 121 bits (303), Expect(2) = 2e-42
Identities = 68/190 (35%), Positives = 106/190 (55%), Gaps = 4/190 (2%)
Frame = +2
Query: 8 VSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMA 67
V+LVL+ V+ D +++AN ++ + G+ ++ SLVGAF+SD+Y R+ TC +F + V
Sbjct: 269 VNLVLFLTRVLGQDNAAAANNVSKWTGTVYIFSLVGAFLSDSYWGRYKTCAIFQGIFVTG 448
Query: 68 LVMITVQAALDNLHPKAC--GKSSCVEGGIAV--MFYTSLCLYALGMGGVRGSLTAFGAN 123
LV ++V L L PK C GK C E MFY S+ L ALG GG + ++ FGA+
Sbjct: 449 LVSLSVTTYLALLRPKGCGNGKLECGEHSSLEMGMFYLSIYLIALGNGGYQPNIATFGAD 628
Query: 124 QFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFV 183
QFDE E+ + FF++ L+ LGS+ T + + + W GF+ ++ + V
Sbjct: 629 QFDEDHSKESYSKVAFFSYFYLALNLGSLFSNTILGYFEDEGLWALGFWASAGSAFLALV 808
Query: 184 TLAIGKPFYR 193
+G P YR
Sbjct: 809 LFLVGTPKYR 838
Score = 108 bits (271), Expect = 4e-24
Identities = 54/171 (31%), Positives = 103/171 (59%), Gaps = 7/171 (4%)
Frame = +2
Query: 355 GVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRKDPSKPISLFWLSFQYAIFGIADMFT 414
G+T+LQR+G+GLV++ I+M AG +E R ++ + +S+FW QYA+ G +++F
Sbjct: 1358 GLTELQRMGIGLVIAIIAMVTAGIVECYRLKYAKQGDTSSLSIFWQIPQYALIGASEVFM 1537
Query: 415 LVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGF 474
VG LEFF + P +KS ++ S+SLG ++S++ V+++ ++ GW+ G
Sbjct: 1538 YVGQLEFFNAQTPDGLKSFGSALCMTSISLGNYVSSLIVSIVMKISTE--DHMPGWIPG- 1708
Query: 475 DLNQNNLNLFYWFLAILSCLNFFNFLYWASWY-------KYKSEEDSNNSK 518
+LN+ +L+ F++ LA+L+ L+ ++ A W+ KY + ++ ++ K
Sbjct: 1709 NLNRGHLDRFFFLLAVLTSLDLIAYIACAKWFQNIQMACKYDNNDEPSSCK 1861
Score = 70.1 bits (170), Expect(2) = 2e-42
Identities = 39/120 (32%), Positives = 63/120 (52%), Gaps = 10/120 (8%)
Frame = +3
Query: 197 PGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDATIE----KIVHTNQMRFLDKAA 252
P +P+ R QV A + +Q+ + + LY + + + KI+HT+ +FLD+AA
Sbjct: 849 PCGNPLSRFCQVFFAASRKLGVQMTSNGDDLYVIDEKESSNNSNRKILHTHGFKFLDRAA 1028
Query: 253 ILQENSESQQ------PWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQG 306
+ Q PW +C +TQVEEVK + R+LPI TI+ + Q+ + V+QG
Sbjct: 1029YITSRDLEVQKGGQHNPWXLCPITQVEEVKCILRLLPIWLCTIIYSVVFTQMASLFVEQG 1208
>TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersicon
esculentum}, partial (38%)
Length = 804
Score = 159 bits (402), Expect = 3e-39
Identities = 86/246 (34%), Positives = 139/246 (55%), Gaps = 6/246 (2%)
Frame = +1
Query: 271 QVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKLGSFTVPASSIPVIPLIF 330
QVEE+KIL RM PI A+ I+ ++ AQ+ T V+QG MN +GSF + +S+ +
Sbjct: 1 QVEELKILIRMFPIWATGIIFSSVYAQMSTLFVEQGTMMNTSIGSFKLSPASLSTFEVAS 180
Query: 331 LCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGR-- 388
+ +P+Y+ VP ++K T G++ QR+G+G +S + M A +E+KR R
Sbjct: 181 VVMWVPVYDKILVPIVKKFTGKKRGISVFQRIGIGPFISGLCMLAAAAVEIKRLQLAREL 360
Query: 389 ----KDPSKPISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSL 444
K P+S+ W QY I G A++FT VG LEFFY E+P M+++ + L+ SL
Sbjct: 361 DLVDKPVGVPLSVLWQIPQYLILGAAEIFTFVGQLEFFYEESPDAMRTICGALPLLNFSL 540
Query: 445 GYFLSTVFVNVINTVTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWAS 504
G +LS+ + ++ T + + GW+ +LN+ +L+ ++ L+ LS LN F+ A
Sbjct: 541 GNYLSSFILTIVTYFTTK--GGRLGWIPD-NLNKGHLDYYFLLLSGLSLLNMLVFIVAAK 711
Query: 505 WYKYKS 510
YK K+
Sbjct: 712 IYKPKN 729
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,447,677
Number of Sequences: 36976
Number of extensions: 278125
Number of successful extensions: 1957
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 1846
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1870
length of query: 569
length of database: 9,014,727
effective HSP length: 101
effective length of query: 468
effective length of database: 5,280,151
effective search space: 2471110668
effective search space used: 2471110668
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148406.6


