

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148403.7 - phase: 0
(315 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
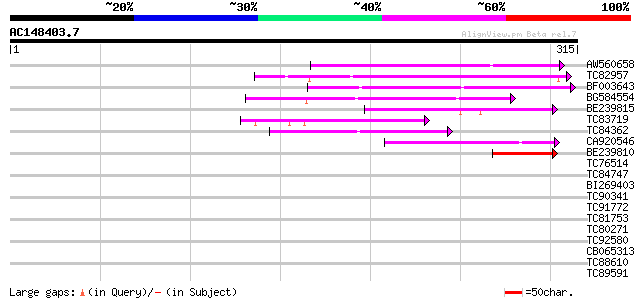
Score E
Sequences producing significant alignments: (bits) Value
AW560658 weakly similar to PIR|T45865|T458 hypothetical protein ... 103 9e-23
TC82957 weakly similar to GP|6957728|gb|AAF32472.1| hypothetical... 75 3e-14
BF003643 weakly similar to PIR|F84686|F846 hypothetical protein ... 74 7e-14
BG584554 72 4e-13
BE239815 69 2e-12
TC83719 similar to GP|18143763|dbj|BAB79812. hypothetical protei... 57 1e-08
TC84362 similar to PIR|T45656|T45656 hypothetical protein F13I12... 53 2e-07
CA920546 48 6e-06
BE239810 44 6e-05
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 30 0.93
TC84747 30 0.93
BI269403 weakly similar to GP|20473411|gb| 36I5.7 {Oryza sativa ... 30 1.6
TC90341 similar to GP|21703099|gb|AAM74492.1 At1g76120/T23E18_5 ... 30 1.6
TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown prot... 29 2.7
TC81753 similar to PIR|G96752|G96752 unknown protein F28P22.2 [i... 28 3.5
TC80271 weakly similar to PIR|T02561|T02561 probable cellulose s... 28 4.6
TC92580 similar to GP|22832529|gb|AAF48893.2 CG7274-PA {Drosophi... 28 4.6
CB065313 similar to PIR|E83237|E8 probable two-component sensor ... 28 4.6
TC88610 homologue to PIR|G96755|G96755 developmental protein hom... 28 6.0
TC89591 similar to SP|Q43093|UGS3_PEA Glycogen [starch] synthase... 28 6.0
>AW560658 weakly similar to PIR|T45865|T458 hypothetical protein F3A4.200 -
Arabidopsis thaliana, partial (6%)
Length = 589
Score = 103 bits (257), Expect = 9e-23
Identities = 56/141 (39%), Positives = 83/141 (58%)
Frame = -1
Query: 168 LPKLVVDDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIIN 227
LP++VVDD + FLN IA+E+ + SF F+D++ID+ DV +L GI++N
Sbjct: 586 LPEIVVDDTSAATFLNXIAYEMCPDFDNDYGICSFAAFMDSLIDNPEDVKLLRSKGILLN 407
Query: 228 YLESDKAVAKLFNSLAKEIPMDREGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAM 287
L SD+ VA+LFN ++ ++ + E E K YC + +W A TYF NPWA+
Sbjct: 406 SLGSDEEVAELFNIISTDLVPNSETYFEVRAKIHDHYCNR-CNTWIAEGFHTYFSNPWAI 230
Query: 288 VSLVAAFFLFALTIIQTIYTV 308
++ +AAF LT IQT +TV
Sbjct: 229 IAFIAAFTALVLTFIQTWFTV 167
>TC82957 weakly similar to GP|6957728|gb|AAF32472.1| hypothetical protein
{Arabidopsis thaliana}, partial (11%)
Length = 857
Score = 75.1 bits (183), Expect = 3e-14
Identities = 51/181 (28%), Positives = 93/181 (51%), Gaps = 5/181 (2%)
Frame = +3
Query: 137 SAMELKEAGISFKKSKTRSLKDVSFNRGV--LRLPKLVVDDHTECMFLNLIAFELLHVDG 194
S EL ++G++F + S+ +SF+ LP + +D +TE NL+A+E V
Sbjct: 186 SVKELVKSGVNFSPTNG-SISSISFDAKTRTFYLPIIGLDVNTEVFLRNLVAYES-SVGS 359
Query: 195 TRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMDREGEL 254
+ + ++ IIDS D IL GII+N+L+SDK VA ++N ++K + + R L
Sbjct: 360 GPLVITRYTELMNGIIDSEDDAKILREKGIILNHLKSDKEVADMWNGMSKSLRLSRVLFL 539
Query: 255 EEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQ---TIYTVGQF 311
++ + + + K +++Y W ++ +AA FL L +Q ++YT +F
Sbjct: 540 DKTIEDVNKFYNSRMKVKMLKFMKSYVFGSWQFLTFLAAIFLLLLMGVQAFCSVYTCSRF 719
Query: 312 Y 312
+
Sbjct: 720 F 722
>BF003643 weakly similar to PIR|F84686|F846 hypothetical protein At2g28580
[imported] - Arabidopsis thaliana, partial (11%)
Length = 545
Score = 73.9 bits (180), Expect = 7e-14
Identities = 42/149 (28%), Positives = 74/149 (49%)
Frame = +3
Query: 166 LRLPKLVVDDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGII 225
L++P+ V TEC+ NLIA E H + + ++V ID +I++ DV +L+ II
Sbjct: 15 LQIPQFKVHQTTECVLRNLIALEQCHYSN-QPFICNYVSLIDFLINTQEDVELLVDKEII 191
Query: 226 INYLESDKAVAKLFNSLAKEIPMDREGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPW 285
++ L S +A + N L K + + + +K++ + WK + L YFR+PW
Sbjct: 192 VHELGSHAELATMINGLCKNVVVTCN-YYGKTSKNLNDHYYNYWKHYMGMLRSVYFRDPW 368
Query: 286 AMVSLVAAFFLFALTIIQTIYTVGQFYQK 314
S + F+F I Q + G ++ +
Sbjct: 369 RFSSTIVGVFIFLFAIAQFLRVTGIYHPR 455
>BG584554
Length = 803
Score = 71.6 bits (174), Expect = 4e-13
Identities = 52/170 (30%), Positives = 82/170 (47%), Gaps = 20/170 (11%)
Frame = +3
Query: 132 DHLNRSAMELKEAGISFKKSKTRSLKDVSFNR--------------------GVLRLPKL 171
+++ R+A +L EAGISF+K + RS D+ F + L++P L
Sbjct: 174 EYVLRTATKLSEAGISFEKLQGRSYCDLKFKKTPILSWFLCLGCVPCFKFVESKLQIPHL 353
Query: 172 VVDDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLES 231
VD TEC+ NLIAFE H + + ++V ID++I + DV +L+ + II + L S
Sbjct: 354 NVDQATECVLRNLIAFEQCHY-SEQPFICNYVSLIDSLIHTHEDVELLVDTEIISHELGS 530
Query: 232 DKAVAKLFNSLAKEIPMDREGELEEVTKSMISYCKKPWKSWRASLIQTYF 281
+A L N L K + + ++ K M + WK + L YF
Sbjct: 531 HAELATLVNGLCKYVVV-TSNCYGKIIKEMNEHYNNWWKHYMGMLRSVYF 677
>BE239815
Length = 586
Score = 69.3 bits (168), Expect = 2e-12
Identities = 40/119 (33%), Positives = 58/119 (48%), Gaps = 12/119 (10%)
Frame = +1
Query: 198 EVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMD---REGEL 254
E SF F+D+++D+ DV L G+ N L SD+ +AKLFN L ++P
Sbjct: 25 ECCSFFTFMDSLVDNGDDVKELRLPGVFQNLLGSDEQLAKLFNDLGDDLPTKMYCNNSYT 204
Query: 255 EEVTKS---------MISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQT 304
+ V S + + WK+W A T+F PWAM++ +AA LT IQT
Sbjct: 205 DAVAYSRRYLLIKVQIEKHYTNKWKTWLAQAYNTHFNTPWAMIAFLAAMLALVLTFIQT 381
>TC83719 similar to GP|18143763|dbj|BAB79812. hypothetical protein
{Clostridium perfringens str. 13}, partial (7%)
Length = 542
Score = 56.6 bits (135), Expect = 1e-08
Identities = 41/115 (35%), Positives = 61/115 (52%), Gaps = 10/115 (8%)
Frame = -1
Query: 129 EDEDHLN----RSAMELKEAGISFKKSKTR--SLKDVSFN----RGVLRLPKLVVDDHTE 178
E+E +LN +S +LK GI +KT + ++SF G LRLP + +D T
Sbjct: 353 EEEINLNWNTYKSIRDLKTVGIQVVANKTDEWNWSNISFTSKWFNGELRLPIFLFNDVTP 174
Query: 179 CMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDK 233
F NL+A+E+ E SF F+D+++D+A DV L +G+ N L SDK
Sbjct: 173 YFFRNLVAYEMCSDVHHTYECCSFFSFMDSLVDNADDVKELRSAGVFQNLLGSDK 9
>TC84362 similar to PIR|T45656|T45656 hypothetical protein F13I12.250 -
Arabidopsis thaliana, partial (5%)
Length = 942
Score = 52.8 bits (125), Expect = 2e-07
Identities = 33/103 (32%), Positives = 52/103 (50%), Gaps = 1/103 (0%)
Frame = +3
Query: 145 GISFK-KSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLIAFELLHVDGTRKEVISFV 203
G+ FK +K+ L D+ F+ VL +P+L V+D TE +F N++A E H + + +V
Sbjct: 585 GVRFKVNTKSECLLDLRFSGRVLEIPQLKVEDWTEILFRNMVALEQCHYP-SESYITDYV 761
Query: 204 CFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEI 246
C I++N+L +VA LFNSL K +
Sbjct: 762 CCFGFPY*YWQGCGYTCSEEILVNWLGDSDSVANLFNSLWKNV 890
>CA920546
Length = 594
Score = 47.8 bits (112), Expect = 6e-06
Identities = 37/99 (37%), Positives = 52/99 (52%), Gaps = 2/99 (2%)
Frame = -2
Query: 209 IIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMD-REGELEEVTKSMISYCKK 267
+I+SA DV+ L G+I + L SD+ VA+L N++AKEI D E L V Y
Sbjct: 590 LINSANDVSALHYKGVIHHSLGSDEHVAELINNIAKEIVPDMNESYLYNVVNEANEYLGC 411
Query: 268 PWKSWRASLIQTYFRNPWAM-VSLVAAFFLFALTIIQTI 305
+RASL+ Y + WA+ +S + A T IQ I
Sbjct: 410 WRARFRASLVHNYLTS-WAVGLSTLGALLALYFTFIQAI 297
>BE239810
Length = 505
Score = 44.3 bits (103), Expect = 6e-05
Identities = 17/36 (47%), Positives = 22/36 (60%)
Frame = +1
Query: 269 WKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQT 304
WK+W A T+F PWAM++ +AA LT IQT
Sbjct: 175 WKTWLAQAYNTHFNTPWAMIAFLAAMLTLVLTFIQT 282
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 30.4 bits (67), Expect = 0.93
Identities = 38/151 (25%), Positives = 61/151 (40%), Gaps = 3/151 (1%)
Frame = +3
Query: 11 ETSETISLEVSEEDEDHLNRSAMELKYTGINFK---SSETISLEVSEEDEDHLNQSAMKL 67
+TSE E S +++ A+ K K S E + E S+EDE+ +A +
Sbjct: 519 DTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPAAKAV 698
Query: 68 KETRINFKSSETSLEVSEEDKDDIIELVIEDHEHALPQSAMVVKKTGINFKTSETSLKVS 127
+ + + E S+ED + E ED + A S V T K + +S + S
Sbjct: 699 PSKNGSVPAKKADTESSDEDSESSDE---EDKKPAAKASKNVSAPTK---KAASSSDEES 860
Query: 128 KEDEDHLNRSAMELKEAGISFKKSKTRSLKD 158
E+ D + K A ++ K K S D
Sbjct: 861 DEESDEDEDAKPVSKPAAVAKKSKKDSSDSD 953
>TC84747
Length = 920
Score = 30.4 bits (67), Expect = 0.93
Identities = 19/67 (28%), Positives = 34/67 (50%)
Frame = -2
Query: 12 TSETISLEVSEEDEDHLNRSAMELKYTGINFKSSETISLEVSEEDEDHLNQSAMKLKETR 71
+ +ISLEV E D ++++K G+ KSS++++ +S L + + R
Sbjct: 802 SDSSISLEVDESSVDKFTGPSLKIKL*GLWLKSSDSVASGISVPSGCRLKKLISL*LKYR 623
Query: 72 INFKSSE 78
I KSS+
Sbjct: 622 IRAKSSD 602
>BI269403 weakly similar to GP|20473411|gb| 36I5.7 {Oryza sativa (japonica
cultivar-group)}, partial (35%)
Length = 505
Score = 29.6 bits (65), Expect = 1.6
Identities = 16/44 (36%), Positives = 21/44 (47%)
Frame = -2
Query: 167 RLPKLVVDDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTII 210
RL KL++ E MFL +D EV F+CF DT +
Sbjct: 408 RLTKLIILQLKETMFLLFRLTNRKTIDNIEVEVPRFICFSDTFL 277
>TC90341 similar to GP|21703099|gb|AAM74492.1 At1g76120/T23E18_5
{Arabidopsis thaliana}, partial (38%)
Length = 1103
Score = 29.6 bits (65), Expect = 1.6
Identities = 15/85 (17%), Positives = 43/85 (49%)
Frame = +2
Query: 4 KYTGINFETSETISLEVSEEDEDHLNRSAMELKYTGINFKSSETISLEVSEEDEDHLNQS 63
K G+ + + LE + + N++ + ++ ++ + + +S+ D++HLN+
Sbjct: 719 KVEGVVGNSKRNLELEAVDVENGSSNKNDV------VDSGVTKDVEVSLSKGDDNHLNKE 880
Query: 64 AMKLKETRINFKSSETSLEVSEEDK 88
++ E +++ + + VS+ED+
Sbjct: 881 SINDNEGKVSVEEVNSKTVVSDEDE 955
>TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown protein
{Arabidopsis thaliana}, partial (17%)
Length = 1237
Score = 28.9 bits (63), Expect = 2.7
Identities = 34/155 (21%), Positives = 65/155 (41%)
Frame = +2
Query: 4 KYTGINFETSETISLEVSEEDEDHLNRSAMELKYTGINFKSSETISLEVSEEDEDHLNQS 63
K G+ + + + S S DE NRS ++ + + E + ++S++ + + S
Sbjct: 377 KAGGLESDAAGSPSPSESNHDE---NRSKKRVRTKKNDSSAKEVAAEDISKKVSEGTSDS 547
Query: 64 AMKLKETRINFKSSETSLEVSEEDKDDIIELVIEDHEHALPQSAMVVKKTGINFKTSETS 123
K+K R + K + D ++ V+ D + + KKT + + +
Sbjct: 548 --KVKPARPSAKKGP----IRSSDVKTVVHAVMADVGSSSLKPEDKKKKTHVKGSSEKGL 709
Query: 124 LKVSKEDEDHLNRSAMELKEAGISFKKSKTRSLKD 158
K S EDED + S++ KS T++ KD
Sbjct: 710 AKSSAEDEDKVTVSSL----------KSATKTTKD 784
>TC81753 similar to PIR|G96752|G96752 unknown protein F28P22.2 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 1273
Score = 28.5 bits (62), Expect = 3.5
Identities = 32/112 (28%), Positives = 48/112 (42%), Gaps = 5/112 (4%)
Frame = +3
Query: 12 TSETISLEVSEEDEDHLNRSAMELKYTGINFKSSETI-SLEVSEE-DEDHLNQS--AMKL 67
++E SL E H +A + K T + ++ + S E S E LN M
Sbjct: 159 SAEFSSLVPEPELNPHRQPAARKKKITSESEVNTAAVTSAEFSSPVPEQELNPQLPVMMT 338
Query: 68 KETRINFKSSETSLEVSE-EDKDDIIELVIEDHEHALPQSAMVVKKTGINFK 118
K R NFK + + + ED D +E + +PQ +M +KTGI K
Sbjct: 339 KGVRGNFKRTNQPKAIEKLEDNDFFVENYLPQ---TVPQRSMAERKTGIPLK 485
>TC80271 weakly similar to PIR|T02561|T02561 probable cellulose synthase
At2g32620 [imported] - Arabidopsis thaliana, partial
(11%)
Length = 553
Score = 28.1 bits (61), Expect = 4.6
Identities = 21/66 (31%), Positives = 30/66 (44%)
Frame = +3
Query: 202 FVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMDREGELEEVTKSM 261
FVC ID + VDV + S I ++Y S+K L + I + E E K
Sbjct: 279 FVCTIDPEKEPTVDVMNTVISAIAMDY-PSNKLSIYLSDDGGSPITLFGIKEAFEFAKVW 455
Query: 262 ISYCKK 267
+ +CKK
Sbjct: 456 VPFCKK 473
>TC92580 similar to GP|22832529|gb|AAF48893.2 CG7274-PA {Drosophila
melanogaster}, partial (2%)
Length = 923
Score = 28.1 bits (61), Expect = 4.6
Identities = 34/133 (25%), Positives = 57/133 (42%)
Frame = +3
Query: 12 TSETISLEVSEEDEDHLNRSAMELKYTGINFKSSETISLEVSEEDEDHLNQSAMKLKETR 71
T ET+++ D+D N ++ + + ++ LE +E D D + M L+ET
Sbjct: 555 TFETVTI-----DDDSTNVGETDM----MKVRQNQDQCLEKAESDCDLV----MLLEET- 692
Query: 72 INFKSSETSLEVSEEDKDDIIELVIEDHEHALPQSAMVVKKTGINFKTSETSLKVSKEDE 131
+ S+E+ + K+ V+ D E +S + + I FK E SK E
Sbjct: 693 -----TPESIEICMDAKE---HRVVSDEEIEDNRSTSSCEASQIPFKEEEVKSIPSKIKE 848
Query: 132 DHLNRSAMELKEA 144
H E+KEA
Sbjct: 849 SHQTSVTKEVKEA 887
>CB065313 similar to PIR|E83237|E8 probable two-component sensor PA3271
[imported] - Pseudomonas aeruginosa (strain PAO1),
partial (17%)
Length = 613
Score = 28.1 bits (61), Expect = 4.6
Identities = 8/22 (36%), Positives = 14/22 (63%)
Frame = -2
Query: 264 YCKKPWKSWRASLIQTYFRNPW 285
+C+ PW+ WR+S+ +PW
Sbjct: 159 WCRPPWR*WRSSVCHGNSTSPW 94
>TC88610 homologue to PIR|G96755|G96755 developmental protein homolog DG1118
[imported] - Arabidopsis thaliana, partial (79%)
Length = 689
Score = 27.7 bits (60), Expect = 6.0
Identities = 26/106 (24%), Positives = 44/106 (40%), Gaps = 4/106 (3%)
Frame = +3
Query: 44 SSETISLEVSEEDEDHLNQSAMKLKETRINFKSS----ETSLEVSEEDKDDIIELVIEDH 99
S T +L+ E D + + + E + F S TSL E + + +++ V +D+
Sbjct: 198 SLATGNLQKMSETMDQFEKQFVNM-EVQAEFMESAMAGSTSLSTPEGEVNSLMQQVADDY 374
Query: 100 EHALPQSAMVVKKTGINFKTSETSLKVSKEDEDHLNRSAMELKEAG 145
+ + V + K + K DED L+R ELK G
Sbjct: 375 GLEVSVNLPVAAAHAVPVKEKDAE----KVDEDDLSRRLAELKARG 500
>TC89591 similar to SP|Q43093|UGS3_PEA Glycogen [starch] synthase
chloroplast precursor (EC 2.4.1.11) (GBSSII), partial
(16%)
Length = 722
Score = 27.7 bits (60), Expect = 6.0
Identities = 27/103 (26%), Positives = 45/103 (43%)
Frame = +1
Query: 65 MKLKETRINFKSSETSLEVSEEDKDDIIELVIEDHEHALPQSAMVVKKTGINFKTSETSL 124
+K K R KS E +ED+DD++ IE + L +++ + +E
Sbjct: 313 LKYKHIRAVGKSIGND-ENEDEDEDDVLNATIEKSKKVLALQKELIQ------QIAERKK 471
Query: 125 KVSKEDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRGVLR 167
VS D D S L+ GIS++ S+ + D + +G R
Sbjct: 472 LVSSIDSD----SIPGLEGNGISYESSEKSASSDSNPQKGSTR 588
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.132 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,850,832
Number of Sequences: 36976
Number of extensions: 92015
Number of successful extensions: 596
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 574
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 590
length of query: 315
length of database: 9,014,727
effective HSP length: 96
effective length of query: 219
effective length of database: 5,465,031
effective search space: 1196841789
effective search space used: 1196841789
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148403.7


